Nomenclature
Short Name:
ACTR2A
Full Name:
Activin receptor type II
Alias:
- Activin A receptor, type IIA
- ACVR2A
- AVR2A
- EC 2.7.11.30
- Kinase ACTR2
- Activin receptor type II precursor
- ACTRII
- ACTRIIA
- ACVR2
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
STKR
SubFamily:
Type2
Specific Links
Structure
Mol. Mass (Da):
57,848
# Amino Acids:
513
# mRNA Isoforms:
2
mRNA Isoforms:
57,848 Da (513 AA; P27037); 45,713 Da (405 AA; P27037-2)
4D Structure:
Interacts with AIP1. Part of a complex consisting of AIP1, ACVR2A, ACVR1B and SMAD3
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
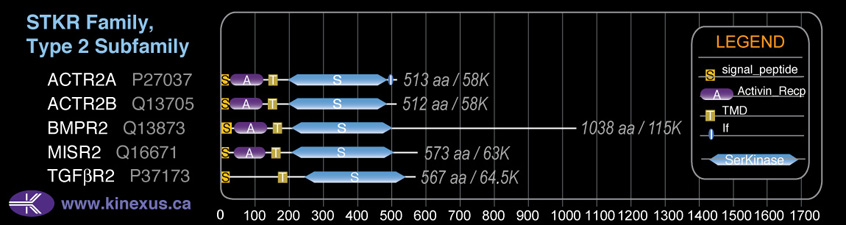
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 19 | signal_peptide |
| 26 | 118 | Activin_recp |
| 139 | 161 | TMD |
| 192 | 485 | Pkinase |
| 486 | 502 | If |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N43, N66.
Serine phosphorylated:
S184.
Threonine phosphorylated:
T486.
Tyrosine phosphorylated:
Y215, Y233.
Ubiquitinated:
K226.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 57
57
1685
22
1865
 2
2
67
11
69
 7
7
220
11
263
 13
13
371
91
610
 32
32
960
24
747
 8
8
248
61
377
 10
10
285
29
478
 21
21
619
39
1023
 25
25
741
10
713
 4
4
120
83
78
 4
4
121
27
138
 21
21
611
125
706
 4
4
127
22
141
 2
2
54
9
33
 4
4
112
24
179
 2
2
53
13
44
 10
10
301
259
2614
 13
13
375
18
884
 5
5
159
71
103
 33
33
982
84
764
 4
4
112
24
145
 3
3
103
26
143
 4
4
111
20
168
 5
5
142
18
178
 3
3
78
24
113
 29
29
875
61
981
 4
4
128
25
143
 6
6
185
18
239
 5
5
160
18
267
 8
8
240
28
230
 25
25
731
18
488
 100
100
2967
26
4912
 3
3
89
53
214
 24
24
720
57
675
 4
4
128
35
113
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 67
67
83.5
100 98.5
98.5
98.5
100 -
-
-
99 -
-
-
99 98.8
98.8
100
99 -
-
-
- 99.6
99.6
100
100 99
99
99.6
99 -
-
-
- 53.8
53.8
60.7
- 92.6
92.6
97.9
93 87.9
87.9
94.9
88 78.8
78.8
89.3
79 -
-
-
- -
-
-
51 53.5
53.5
69.7
- 25.3
25.3
40.5
- 51.4
51.4
64.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
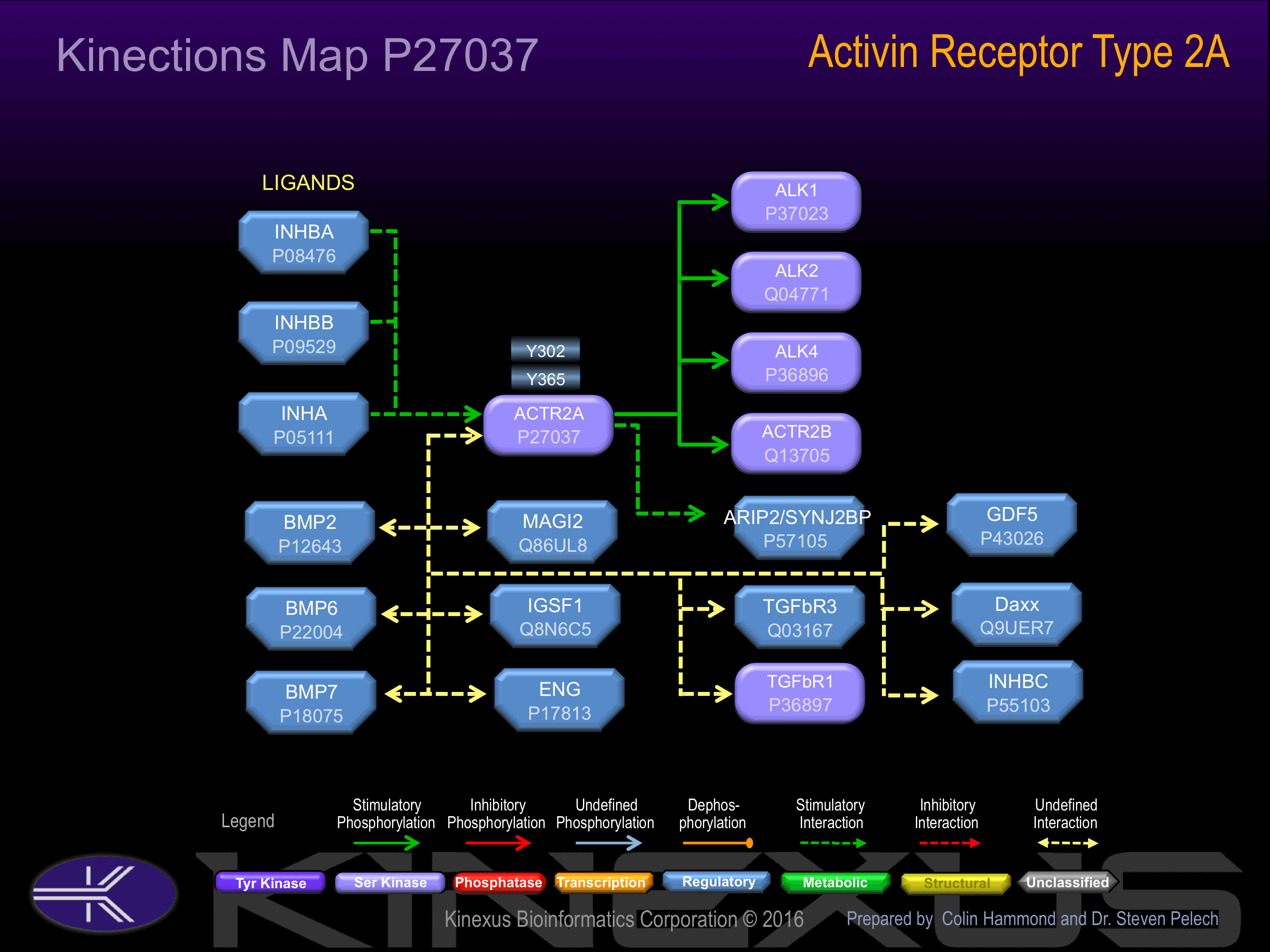
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | INHBA - P08476 |
| 2 | ACVRL1 - P37023 |
| 3 | BMP7 - P18075 |
| 4 | SYNJ2BP - P57105 |
| 5 | ACVR1B - P36896 |
| 6 | INHBB - P09529 |
| 7 | BMP6 - P22004 |
| 8 | DAXX - Q9UER7 |
| 9 | TGFBR3 - Q03167 |
| 10 | INHA - P05111 |
| 11 | ACVR1 - Q04771 |
| 12 | GDF5 - P43026 |
| 13 | INHBC - P55103 |
| 14 | BMP2 - P12643 |
| 15 | TCTEX1D4 - Q5JR98 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
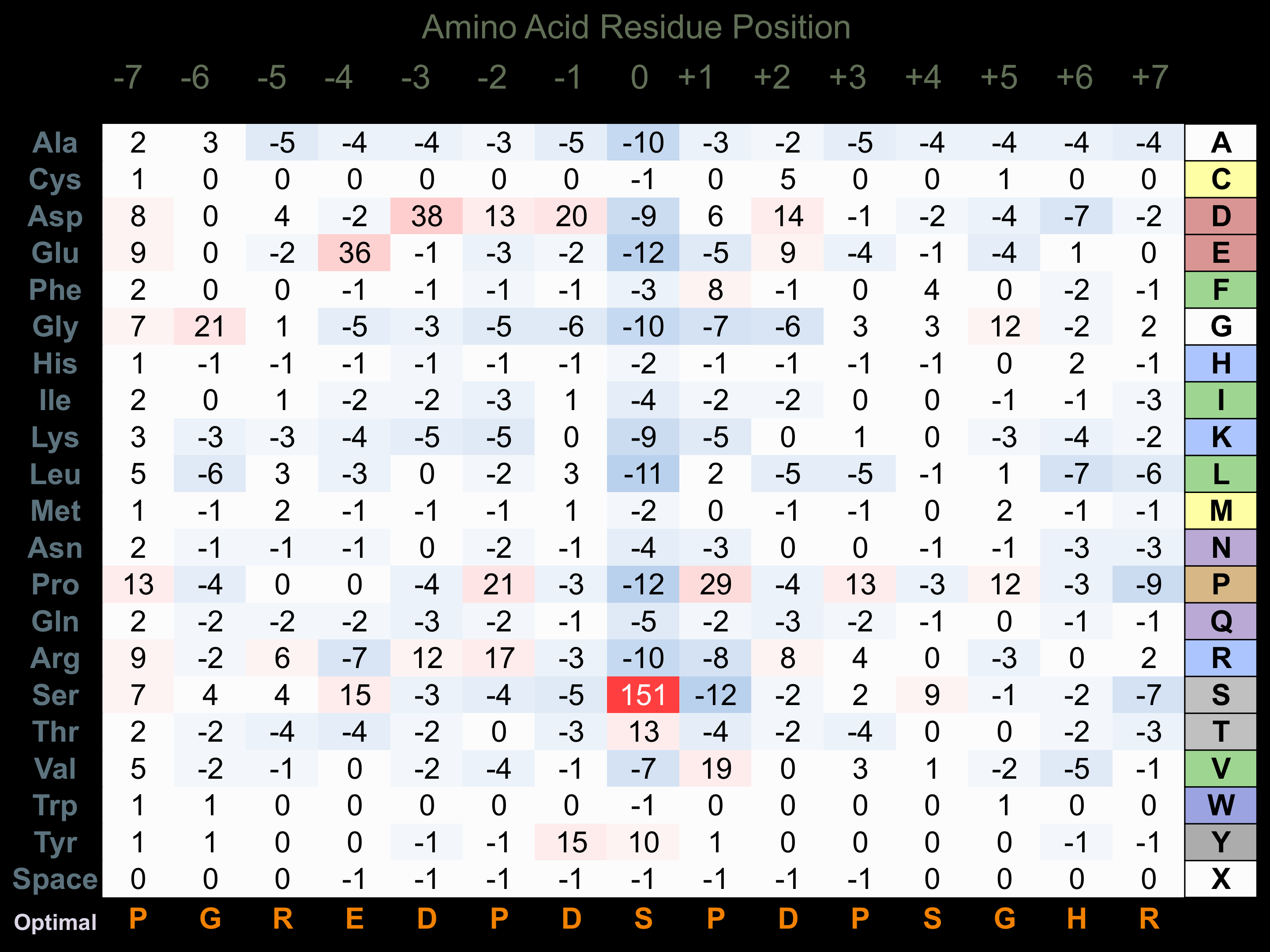
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| PD173955 | Kd = 10 nM | 447077 | 386051 | 22037378 |
| PP242 | Kd = 18 nM | 25243800 | 22037378 | |
| Dasatinib | Kd = 210 nM | 11153014 | 1421 | 18183025 |
| KW2449 | Kd = 320 nM | 11427553 | 1908397 | 22037378 |
| CHEMBL249097 | Kd < 400 nM | 25138012 | 249097 | 19035792 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| AC1O6ZUA | Kd < 1.25 µM | 6539569 | 408019 | 19035792 |
| CHEMBL511337 | Kd < 1.25 µM | 44588220 | 511337 | 19035792 |
| NVP-TAE684 | Kd = 2.2 µM | 16038120 | 509032 | 22037378 |
| Lestaurtinib | Kd = 2.5 µM | 126565 | 22037378 | |
| JNJ-7706621 | Kd = 2.9 µM | 5330790 | 191003 | 18183025 |
| Alvocidib | Kd = 3.1 µM | 9910986 | 428690 | 22037378 |
Disease Linkage
General Disease Association:
Dystostosis
Specific Diseases (Non-cancerous):
Multiple Synostoses Syndrome
Comments:
Multiple Synostoses Syndrome is a dysostosis that is characterized by premature joint ankylosis and may involve autosomal dominant inheritance.
Comments:
ACTR2A appears to be a tumour suppressor protein (TSP). Cancer-related mutations in human tumours point to a loss of function of the protein kinase. The active form of the protein kinase normally acts to inhibit cell proliferation, except in fibroblasts where TGF-beta can stimulate their growth.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Pituitary adenomas (ACTH-secreting) (%CFC= -65); Skin fibrosarcomas (%CFC= -52); Skin melanomas (%CFC= -63, p<0.009); and Skin melanomas - malignant (%CFC= -53, p<0.003). The COSMIC website notes an up-regulated expression score for ACTR2A in diverse human cancers of 394, which is 0.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 50 for this protein kinase in human cancers was 0.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.21 % in 25478 diverse cancer specimens. This rate is 2.7-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None >4 in 20199 cancer specimens
Comments:
Eighty K437fs*5 deletions frame shift mutants and three R438fs*19 insertions frame shift mutants listed on the COSMIC website. These results in truncation of the C-terminal portion of the kinase catalytic domain of ACTR2A.