Nomenclature
Short Name:
ALK
Full Name:
ALK tyrosine kinase receptor
Alias:
- Anaplastic lymphoma kinase
- Kinase ALK
- Anaplastic lymphoma kinase (Ki-1)
- Anaplastic lymphoma receptor tyrosine kinase
- CD246
- EC 2.7.10.1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Alk
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
176442
# Amino Acids:
1620
# mRNA Isoforms:
1
mRNA Isoforms:
176,442 Da (1620 AA; Q9UM73)
4D Structure:
Homodimer. When bound to ligand.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
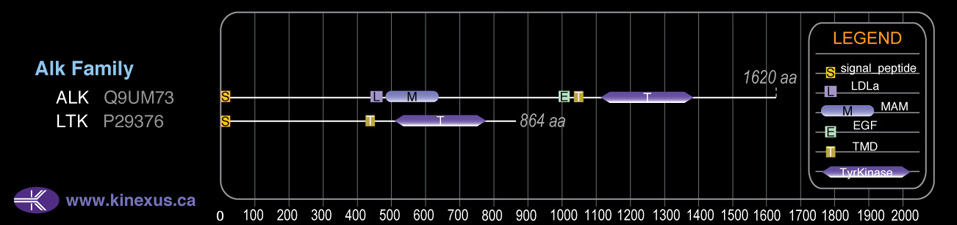
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
Predicted to be N-linked GlcNAcylated at N169, N244, N285, N324, N411, N424, N445, N563, N571, N627, N709, N808, N863, N864, N886, N986.
Serine phosphorylated:
S45, S47, S62, S211, S312, S319, S326, S329, S1075, S1104, S1481, S1495, S1509.
Threonine phosphorylated:
T1087, T1090, T1506, T1512, T1597, T1607.
Tyrosine phosphorylated:
Y46, Y1078, Y1092, Y1096, Y1131, Y1278+, Y1282+, Y1283+, Y1359, Y1401, Y1507, Y1584, Y1586, Y1604.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 78
78
1021
29
1418
 2
2
20
14
25
 13
13
174
27
601
 21
21
272
101
573
 36
36
472
25
456
 0.8
0.8
10
74
6
 20
20
263
35
470
 35
35
452
55
809
 23
23
297
17
250
 6
6
84
107
350
 10
10
135
50
579
 37
37
483
198
585
 11
11
146
49
640
 1
1
18
10
15
 17
17
219
40
772
 3
3
42
15
57
 3
3
35
288
229
 11
11
149
36
502
 5
5
59
106
246
 22
22
284
109
324
 11
11
138
42
493
 13
13
173
46
681
 14
14
178
34
590
 14
14
178
35
608
 14
14
179
41
680
 30
30
385
67
554
 15
15
196
52
950
 26
26
333
36
1200
 14
14
188
31
643
 10
10
128
28
99
 22
22
289
24
282
 100
100
1304
36
4284
 0.5
0.5
6
34
7
 53
53
696
57
667
 24
24
313
35
300
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 32.2
32.2
39.6
97 75.8
75.8
77
- -
-
-
90.6 -
-
-
- 78.3
78.3
81
92.5 -
-
-
- 87.2
87.2
90.4
88 88.1
88.1
90.9
89 -
-
-
- -
-
-
- 69.8
69.8
81.1
74 20.1
20.1
35.1
62 42.2
42.2
54.9
61 -
-
-
- 28.3
28.3
42.8
34 31.4
31.4
46.6
- -
-
-
32 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
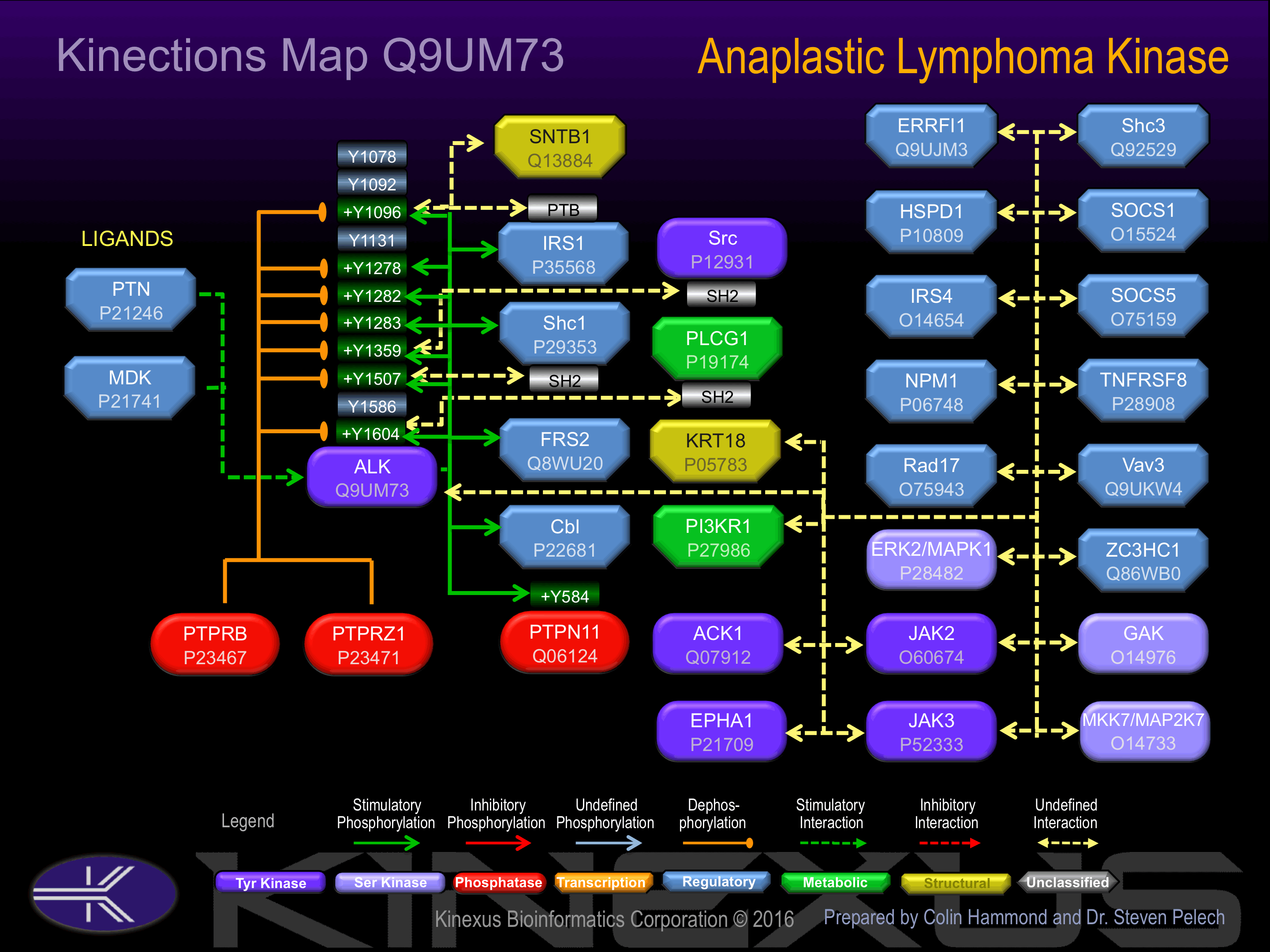
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PTPRZ1 - P23471 |
| 2 | TNFRSF8 - P28908 |
| 3 | NPM1 - P06748 |
| 4 | IRS1 - P35568 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| PTPN11 (SHP2) | Q06124 | Y584 | REDSARVYENVGLMQ | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
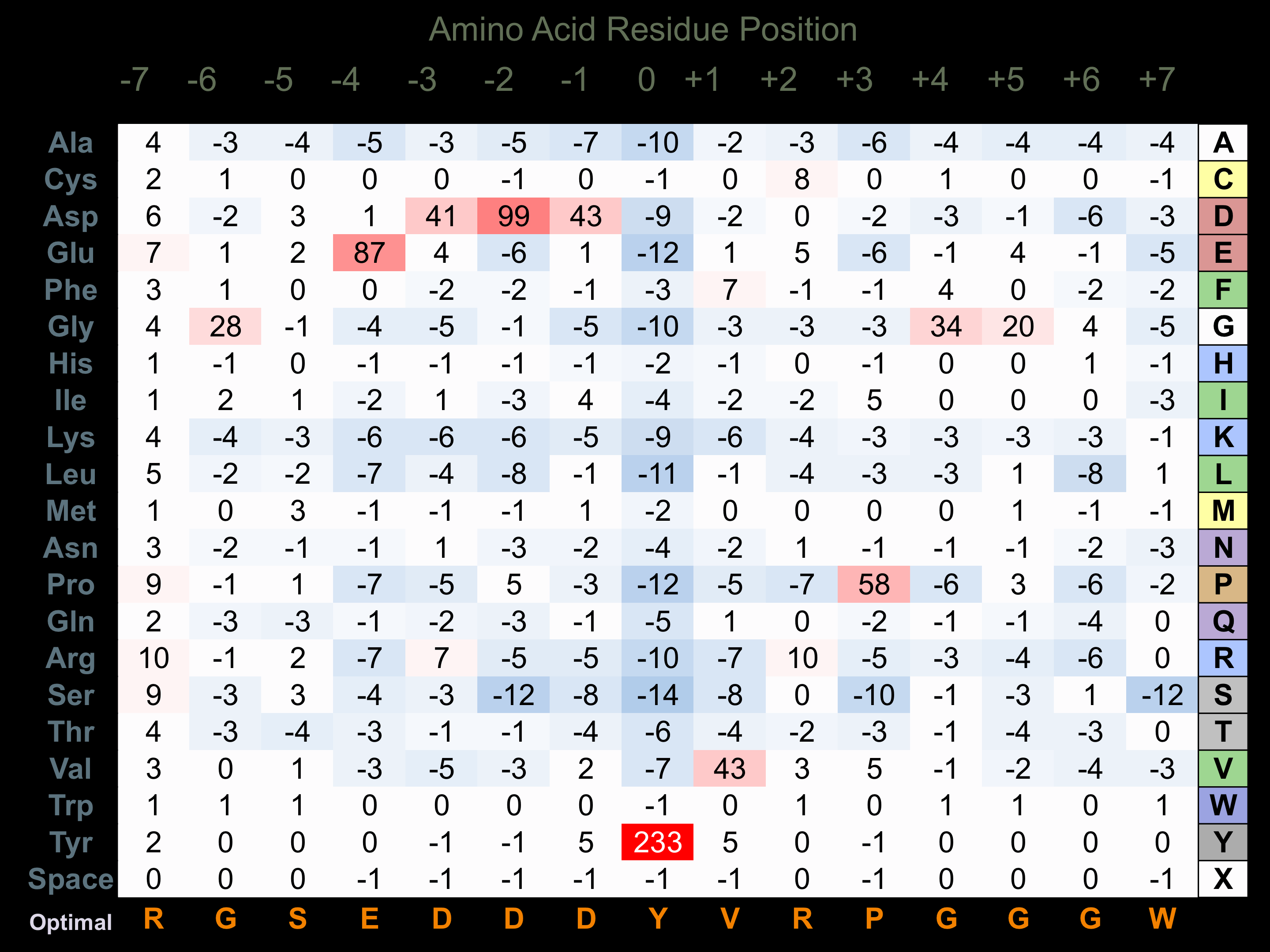
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
ALK-negative anaplastic large cell lymphomas; ALK-positive anaplastic large cell lymphomas; ALK-related neuroblastomas susceptibility; ALK-positive large B-cell lymphomas; ALK+ histiocytosis; neuroblastomas; Anaplastic large cell lymphomas (ALCL); Inflammatory myofibroblastic tumours (IMTs); lung cancer; neuroblastomas 3; Reticulosarcomas; neuroblastomas; Susceptibility; Primary cutaneous anaplastic large cell lymphomas; Follicular dendritic Cell tumours; Tracheal lymphomas
Comments:
ALK is a known oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. Chromosomal rearrangments (most common), mutations, and amplifications with the ALK gene are associated with numerous tumours including: anaplastic large cell lymphomas, neuroblastomas, and non-small cell lung cancer. These fusion proteins, as well as other mutations and amplifications, likely confer heightened (possibly consititutive) phosphotransferase activity and promote cell growth and anti-apoptotic pathways such as the Akt and MAPK pathways. In Neuroblastoma 3 (NBLST3), there was constitutive activation, and Endoplasmic Reticulum or Golgi Apparatus localization when ALK had gain of function mutations with F1174I or F1174V (located in the kinase catalytic subdomain III) or R1275Q (located just before the kinase catalytic Subdomain VII). NBLST3 is a common neoplasm of early childhood resulting from the embryonic cells that form the primitive neural crest and generate the adrenal medulla and the sympathetic nervous system. Large-cell lymphomas make up 25% of non-Hodgkin lymphomas. Non-Hodgkin lymphomas are cancers beginning in the lymphatic system, and they arise from B- or T- cells. Non-Hodgkin lymphomas spread in an unpredictable way (distinguishing it from Hodgkin lymphomas), and manifests in at least 30 forms of cancer. Translocations of the ALK gene resulting in fusion proteins with NPM1 are observed in 5-10% of non-Hodgkin lymphomas, with CARS and SEC31A in inflammatory myofibroblastic tumours (IMTs) and ALO17 in anaplastic large-cell lymphoma (ALCL). The genes C/EBPB, and BCL2A1 are essential for ALCL growth and transcriptionally induced by ALK.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -53, p<0.0001); Breast epithelial cell carcinomas (%CFC= -47, p<0.061); and Ovary adenocarcinomas (%CFC= +169, p<0.096). The COSMIC website notes an up-regulated expression score for ALK in diverse human cancers of 353, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.16 % in 32013 diverse cancer specimens. This rate is 2.2-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.57 % in 1279 large intestine cancers tested; 0.55 % in 2710 autonomic ganglia cancers tested; 0.52 % in 994 skin cancers tested; 0.27 % in 2128 lung cancers tested; 0.25 % in 619 stomach cancers tested; 0.2 % in 603 endometrium cancers tested; 0.17 % in 607 oesophagus cancers tested; 0.11 % in 694 thyroid cancers tested; 0.1 % in 930 upper aerodigestive tract cancers tested; 0.08 % in 968 ovary cancers tested; 0.06 % in 2042 haematopoietic and lymphoid cancers tested; 0.06 % in 1271 liver cancers tested; 0.05 % in 2315 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: F1174L (146); F1174C (13); F1174V (7); F1174I (6); R1275Q (87); F125V (11); F125C (9); F125I (6).
Comments:
Only 7 deletions, 2 insertions and 2 complex mutations are noted on the COSMIC website.