Nomenclature
Short Name:
AurB
Full Name:
Serine-threonine-protein kinase 12
Alias:
- AIK2
- Aurora,IPL1-related kinase 2
- Aurora-2
- Aurora-B
- EC 2.7.11.1
- STK12
- AIM1
- AIM-1
- ARK2
- AURKB
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
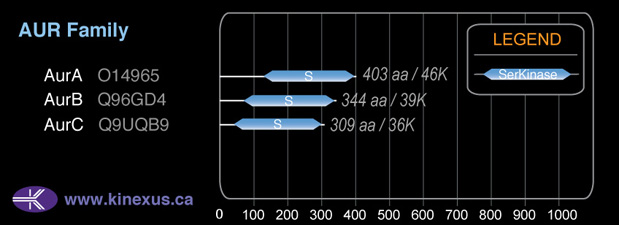
Family:
AUR
SubFamily:
NA
Structure
Mol. Mass (Da):
39,280
# Amino Acids:
344
# mRNA Isoforms:
5
mRNA Isoforms:
39,467 Da (345 AA; Q96GD4-5); 39,311 Da (344 AA; Q96GD4); 35,302 Da (312 AA; Q96GD4-2); 34,760 Da (303 AA; Q96GD4-4); 16,211 Da (142 AA; Q96GD4-3)
4D Structure:
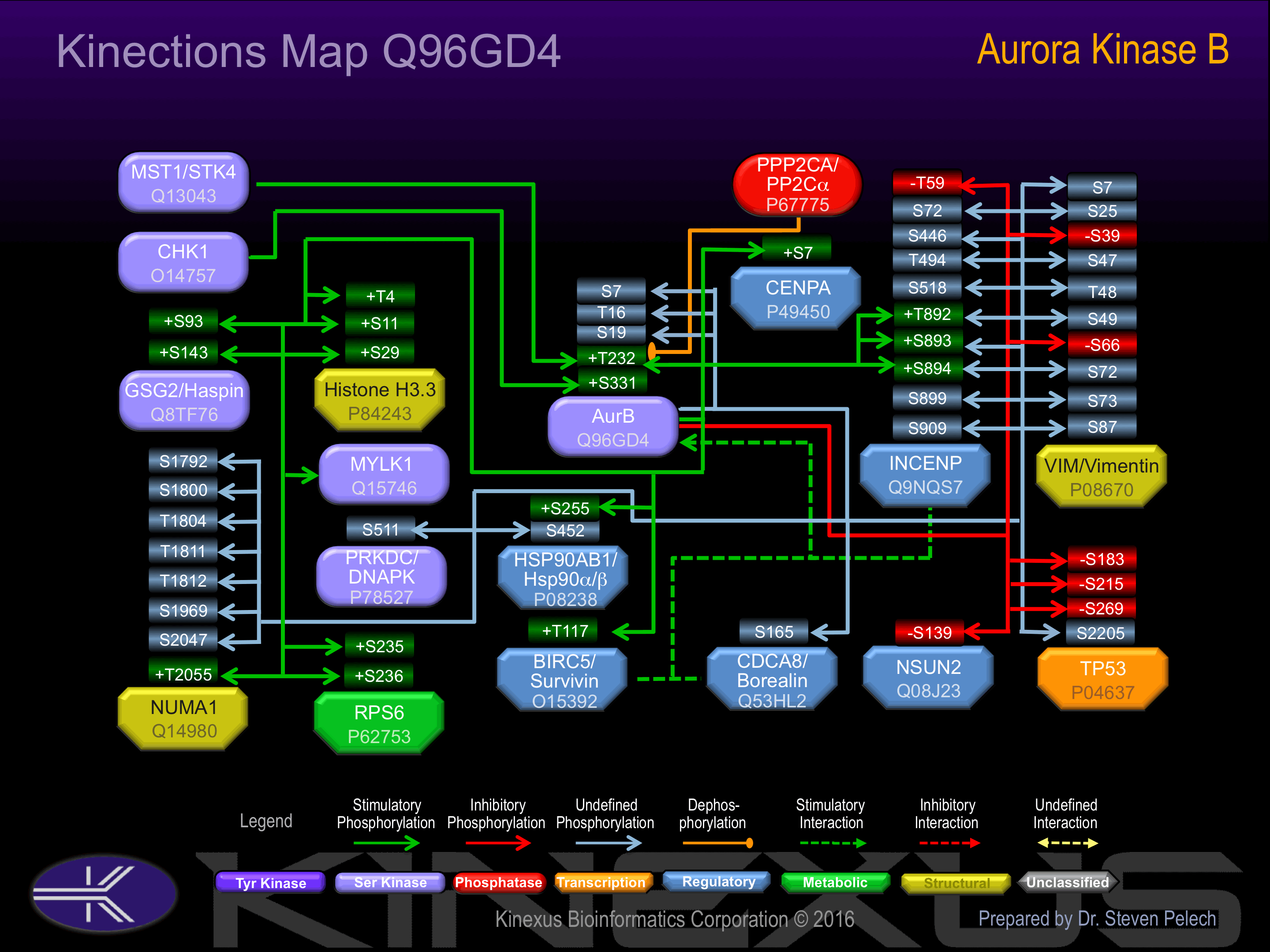
Interacts with TACC1. Associates with RACGAP1 during M phase. Component of the CPC at least composed of BIRC5/survivin CDCA8/borealin, INCENP and AURKB/Aurora-B. Interacts with CDCA1 and NDC80. Interacts with EVI5. Interacts with SEPT1. Interacts with PSMA3
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 77 | 327 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2.
Serine phosphorylated:
S7, S45, S61, S62, S227, S313, S331.
Sumoylated:
K85, K87, K202.
Threonine phosphorylated:
T16, T35, T64, T73, T232, T236+.
Tyrosine phosphorylated:
Y8, Y12, Y92, Y239.
Ubiquitinated:
K56, K85, K87, K115, K164, K168, K202, K211, K215, K287, K291, K306.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 55
55
2072
22
3215
 0.4
0.4
16
10
20
 31
31
1151
9
1551
 47
47
1756
135
4585
 13
13
483
19
327
 10
10
371
63
758
 7
7
249
29
559
 52
52
1945
34
4662
 8
8
309
10
232
 4
4
164
79
465
 91
91
3448
63
5314
 14
14
517
111
601
 16
16
605
20
1629
 1.1
1.1
40
9
37
 15
15
564
21
1157
 0.3
0.3
10
13
22
 11
11
396
174
2722
 21
21
793
15
1731
 60
60
2279
121
5278
 10
10
379
84
370
 11
11
433
20
891
 13
13
499
22
1532
 24
24
893
10
1511
 100
100
3771
68
6378
 25
25
960
20
1207
 51
51
1942
55
5219
 10
10
378
23
757
 67
67
2529
16
7254
 16
16
622
16
1628
 2
2
90
28
62
 19
19
703
18
503
 23
23
851
26
1531
 21
21
790
64
1121
 20
20
770
52
664
 3
3
98
35
49
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 85.8
85.8
86.3
98 60.3
60.3
60.7
98 -
-
-
93 -
-
-
- 91.3
91.3
95.1
92 -
-
-
- 84.9
84.9
91.6
85 84.6
84.6
90.4
85 -
-
-
- 61.9
61.9
72.1
- 57
57
66.5
- 67.6
67.6
78.7
71 66.6
66.6
76.2
75 -
-
-
- 44.5
44.5
62.5
- -
-
-
- 53.5
53.5
65.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
65 54.9
54.9
68
61 36.8
36.8
59.1
44.5 -
-
-
58
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BIRC5 - O15392 |
| 2 | INCENP - Q9NQS7 |
| 3 | RACGAP1 - Q9H0H5 |
| 4 | CDCA8 - Q53HL2 |
| 5 | C2orf18 - Q8N357 |
| 6 | 38960 - Q8WYJ6 |
| 7 | MAPRE1 - Q15691 |
| 8 | TACC1 - O75410 |
| 9 | SGOL1 - Q5FBB7 |
| 10 | HIST1H3A - P68431 |
| 11 | NSUN2 - Q08J23 |
| 12 | PSMA3 - P25788 |
| 13 | RCC2 - Q9P258 |
| 14 | BARD1 - Q99728 |
| 15 | BRCA2 - P51587 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AHNAK | Q09666 | T158 | ITVTRRVTAYTVDVT | |
| AurB (STK12) | Q96GD4 | S19 | YGRQTAPSGLSTLPQ | |
| AurB (STK12) | Q96GD4 | S7 | _MAQKENSYPWPYGR | |
| AurB (STK12) | Q96GD4 | T16 | PWPYGRQTAPSGLST | |
| AurB (STK12) | Q96GD4 | T232 | APSLRRKTMCGTLDY | + |
| BIRC5 (Survivin) | O15392 | T117 | KNKIAKETNNKKKEF | + |
| C19orf21 | Q8IVT2 | S348 | RRERGRPSLYVQRDI | |
| CBX5 | P45973 | S92 | SESNKRKSNFSNSAD | |
| CCDC86 | Q9H6F5 | S255 | DRSKKRFSQMLQDkP | ? |
| CDCA5 | Q96FF9 | S83 | PRRSPRISFFLEKEN | |
| CDCA8 (Borealin) | Q53HL2 | S165 | KGKGKRSSRANTVTP | |
| CENPA | P49450 | S7 | _MGPRRRSRKPEAPR | + |
| CKAP2 | Q8WWK9 | T39 | EHLLRRkTLFAYKQE | ? |
| COBLL1 | Q53SF7 | S993 | SPEQTLSPLSKMPHS | |
| DDA1 | Q9BW61 | S33 | KASNRRPSVYLPTRE | |
| DDX3X | O00571 | S594 | RSKSSrFSGGFGARD | |
| DDX3X | O00571 | S605 | GARDYRQSSGASSSS | |
| DDX3X | O00571 | S82 | RSDSRGKSSFFSDrG | ? |
| DDX52 | Q9Y2R4 | S22 | KFDTRRFSADAARFQ | |
| DEK | P35659 | T67 | KKKVERLTMQVSSLQ | |
| Desmin | P17661 | S11 | YSSSQRVSSYRRTFG | |
| Desmin | P17661 | S59 | VYQVSRTSGGAGGLG | |
| Desmin | P17661 | T16 | RVSSYRRTFGGAPGF | |
| DHX8 | Q14562 | S1214 | EPNAWRISRAFRRR_ | |
| DSN1 | Q9H410 | S100 | RQSWRRASMKETNRR | |
| DSN1 | Q9H410 | S109 | KETNRRKSLHPIHQG | |
| GFAP | P14136 | S13 | ITSAARRSYVSSGEM | |
| GFAP | P14136 | S38 | LGPGTRLSLARMPPP | |
| GFAP | P14136 | T7 | _MERRRITSAARRSY | |
| GSG2/Haspin | Q8TF76 | S143 | PPFPSRDSGRLSPDL | + |
| GSG2/Haspin | Q8TF76 | S93 | RVPKDRPSLTVTPKR | + |
| H3.1 | P68431 | S11 | TKQTARKSTGGKAPR | + |
| H3.1 | P68431 | S29 | ATKAARKSAPATGGV | + |
| H3.1 | P68431 | T4 | ____MARTKQTARKS | + |
| HEC1 | O14777 | S15 | SGGAGRLSMQELRSQ | |
| HEC1 | O14777 | S5 | ___MKRSSVSSGGAG | |
| HEC1 | O14777 | S55 | PTSERKVSLFGKRTS | |
| HEC1 | O14777 | S69 | SGHGSRNSQLGIFSS | |
| HEC1 | O14777 | T49 | KLSINKPTSERKVSL | |
| HIST1H2BJ | P06899 | S79 | ERIAGEASRLAHYNK | |
| HIST1H2BJ | P06899 | Y84 | EASRLAHYNKRSTIT | |
| HMGN2 | P05204 | S25 | KDEPQRRSARLSAKP | ? |
| HMGN2 | P05204 | S29 | QRRSARLSAkPAPPk | |
| HMGN5 | P82970 | S20 | RQEPKRRSARLSAML | |
| HMGN5 | P82970 | S24 | KRRSARLSAMLVPVT | |
| hnRNPA1 | P09651 | S337 | GGNFGGrSSGPYGGG | |
| hnRNPA1 | P09651 | S338 | GNFGGrSSGPYGGGG | |
| HSP90AB1 | P08238 | S255 | PKIEDVGSDEEDDSG | + |
| HSP90AB1 | P08238 | S452 | STNRRRLSELLRYHT | |
| INCENP | Q9NQS7 | S446 | GPREPPQSARRKRSY | |
| INCENP | Q9NQS7 | S518 | APRSVMKSFIKRNTP | |
| INCENP | Q9NQS7 | S72 | RRKKRRISYVQDENR | |
| INCENP | Q9NQS7 | S893 | PRYHKRTSSAVWNSP | + |
| INCENP | Q9NQS7 | S894 | RYHKRTSSAVWNSPP | + |
| INCENP | Q9NQS7 | S909 | LQGARVPSSLAYSLK | |
| INCENP | Q9NQS7 | T494 | kVVRPLRTFLHTVQR | |
| INCENP | Q9NQS7 | T59 | EPELMPKTPSQKNRR | - |
| INCENP | Q9NQS7 | T892 | KPRYHKRTSSAVWNS | + |
| KIF23 | Q02241 | S911 | NGSRKRRSSTVAPAQ | |
| KIF2C | Q99661 | S109 | APKESLRSRSTRMST | |
| KIF2C | Q99661 | S111 | KESLRSRSTRMSTVS | |
| KIF2C | Q99661 | S115 | RSRSTRMSTVSELRI | |
| KIF2C | Q99661 | S192 | VNSVRRKSCLVKEVE | |
| KIF2C | Q99661 | S95 | IQKQKRRSVNSKIPA | |
| KRT18 | P05783 | S7 | _MSFTTRSTFSTNYR | |
| KRT18 | P05783 | T8 | MSFTTRSTFSTNYRS | |
| KRT7 | P08729 | S27 | RGAQVRLSSARPGGL | |
| KRT8 | P05787 | S34 | SGPGSrISSSSFSRV | |
| KRT8 | P05787 | T6 | __MSIRVTQkSYkVS | |
| LMO7 | Q8WWI1 | S116 | IKKINRLSTPIAGLD | |
| LMO7 | Q8WWI1 | S1197 | LKNLKRRSQFFEQGS | |
| LMO7 | Q8WWI1 | S1204 | SQFFEQGSSDSVVPD | |
| LMO7 | Q8WWI1 | S1205 | QFFEQGSSDSVVPDL | |
| LMO7 | Q8WWI1 | S1207 | FEQGSSDSVVPDLPV | |
| LMO7 | Q8WWI1 | S300 | DMSYRRISAVEPKTA | |
| LMO7 | Q8WWI1 | S751 | KWKDRRKSYTSDLQK | |
| LUZP1 | Q86V48 | S877 | PETVVSRSSIIIKPS | |
| LUZP1 | Q86V48 | S878 | ETVVSRSSIIIKPSD | |
| MgcRacGAP (RACGAP1) | Q9H0H5 | S387 | ETGLYRISGCDRTVK | |
| MgcRacGAP (RACGAP1) | Q9H0H5 | S410 | VKTVPLLSKVDDIHA | |
| MgcRacGAP (RACGAP1) | Q9H0H5 | T249 | WTRSRRKTGTLQPWN | |
| MKI67 | P46013 | S374 | YTTGRRESVNLGkSE | |
| MKI67 | P46013 | S538 | EAPTKRkSLVMHTPP | |
| MPHOSPHO10 | O00566 | T332 | KESLKRVTFALPDDA | |
| MRLC2 (MYL12B) | P19105 | S20 | KRPQRATSNVFAMFD | |
| MSH6 | P52701 | S5 | ___MSRQSTLYSFFP | |
| MYBBP1A | Q9BQG0 | S1303 | ARKKARLSLVIRSPS | |
| NAGK | Q9UJ70 | S76 | RSLGLSLSGGDQEDA | |
| NSUN2 | Q08J23 | S139 | SRKILRKSPHLEKFH | - |
| NUMA1 | Q14980 | S1169 | QETLRRASMQPIQIA | |
| NUMA1 | Q14980 | S1792 | PLESSLDSLGDVFLD | |
| NUMA1 | Q14980 | S1800 | LGDVFLDSGRKTRSA | |
| NUMA1 | Q14980 | S2047 | KQADRRQSMAFSILN | |
| NUMA1 | Q14980 | T1804 | FLDSGRKTRSARRRT | |
| NUMA1 | Q14980 | T1811 | TRSARRRTTQIINIT | |
| NUMA1 | Q14980 | T1812 | RSARRRTTQIINITM | |
| NUMA1 | Q14980 | T2055 | MAFSILNTPkKLGNS | + |
| NUSAP1 | Q9BXS6 | S240 | VPPRGRLSVASTPIS | |
| p53 | P04637 | S183 | CPHHERCSDSDGLAP | - |
| p53 | P04637 | S215 | DRNTFRHSVVVPYEP | - |
| p53 | P04637 | S269 | GNLLGRNSFEVRVCA | - |
| p53 | P04637 | S284 | CPGRDRRTEEENLRK | |
| PAI-RBP1 | Q8NC51 | S199 | EFDRHSGSDrSSFSH | |
| PAI-RBP1 | Q8NC51 | S202 | RHSGSDrSSFSHYSG | |
| PAI-RBP1 | Q8NC51 | S203 | HSGSDrSSFSHYSGL | |
| PAI-RBP1 | Q8NC51 | S205 | GSDrSSFSHYSGLkH | |
| PLEC1 | Q15149 | T2886 | PVRNRRLTVNEAVKE | |
| PPHLN1 | Q8NEY8 | S110 | RDGFRRKSFYSSHYA | |
| PRKDC | P78527 | S511 | ESEDHRASGEVRTGK | |
| PRRC2C (BAT2D1) | Q9Y520 | S1013 | NDRPVRRSGPIKKPV | |
| PTRF | Q6NZI2 | S300 | SRDKLRkSFTPDHVV | |
| RASSF1 | Q9NS23 | S207 | TSVRRRTSFYLPKDA | |
| RBM14 | Q96PK6 | S618 | LSDYRRLSESQLSFR | |
| RBM14 | Q96PK6 | S649 | HSDYARYSGSYNDYL | |
| RBM3 | P98179 | S147 | QGGYDRYSGGNYRDN | |
| RBMX | P38159 | S189 | PVSRGRDSYGGPPRR | |
| RBMX | P38159 | S326 | GYGGSRDSYSSSRSD | |
| RDBP | P18615 | S131 | SRRPQRkSLYESFVS | |
| RIF1 | Q5UIP0 | S2205 | VNKVRRVSFADPIYQ | |
| RND3 | P61587 | S218 | QRATKRISHMPSRPE | |
| RND3 | P61587 | S222 | KRISHMPSRPELSAV | |
| RPL21 | P46778 | S104 | KHSKSRDSFLKRVKE | |
| RPL8 | P62917 | S130 | RGKLARASGNYATVI | |
| RPS10 | P46783 | T118 | GERPARLTRGEADRD | |
| RPS6 | P62753 | S235 | IAKRRRLSSLRASTS | + |
| RPS6 | P62753 | S236 | AKRRRLSSLRASTSK | + |
| RSL1D1 | O76021 | T312 | KRQQARKTASVLSKD | |
| SEPT1. | Q8WYJ6 | S248 | PVRGRRYSWGTVEVE | |
| SEPT1. | Q8WYJ6 | S307 | PGARDRASRSKLSRQ | |
| SEPT1. | Q8WYJ6 | S315 | RSKLSRQSATEIPLP | |
| SF3B5 | Q9BWJ5 | T6 | __MTDRYTIHSQLEH | |
| SF3B5 | Q9BWJ5 | Y5 | ___MTDRYTIHSQLE | |
| SH2D4A | Q9H788 | S261 | REDYKRLSLGAQKGR | |
| SLTM | Q9NWH9 | T855 | GERDERRTVIIHDRP | |
| SSH3 | Q8TE77 | S37 | SRLQRRQSFAVLRGA | |
| SUPT16H | Q9Y5B9 | T494 | EEAKRRLTEQKGEQQ | |
| UTP3 | Q9NQZ2 | S462 | RKEEQRYSGELSGIR | |
| Vimentin | P08670 | S25 | PGTASRPSSSRSYVT | |
| Vimentin | P08670 | S39 | TTSTRTYSLGSALRP | - |
| Vimentin | P08670 | S47 | LGSALRPSTSRSLYA | |
| Vimentin | P08670 | S49 | SALRPSTSRSLYASS | |
| Vimentin | P08670 | S65 | GGVYATRSSAVRLRS | |
| Vimentin | P08670 | S66 | GVYATRSSAVRLRSS | - |
| Vimentin | P08670 | S7 | _MSTRSVSSSSYRRM | |
| Vimentin | P08670 | S72 | SSAVRLRSSVPGVRL | |
| Vimentin | P08670 | S73 | SAVRLRSSVPGVRLL | |
| Vimentin | P08670 | S87 | LQDSVDFSLADAINT | |
| Vimentin | P08670 | T48 | GSALRPSTSRSLYAS | |
| ZC3H11A | O75152 | S758 | SMKTRRLSSASTGKP |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
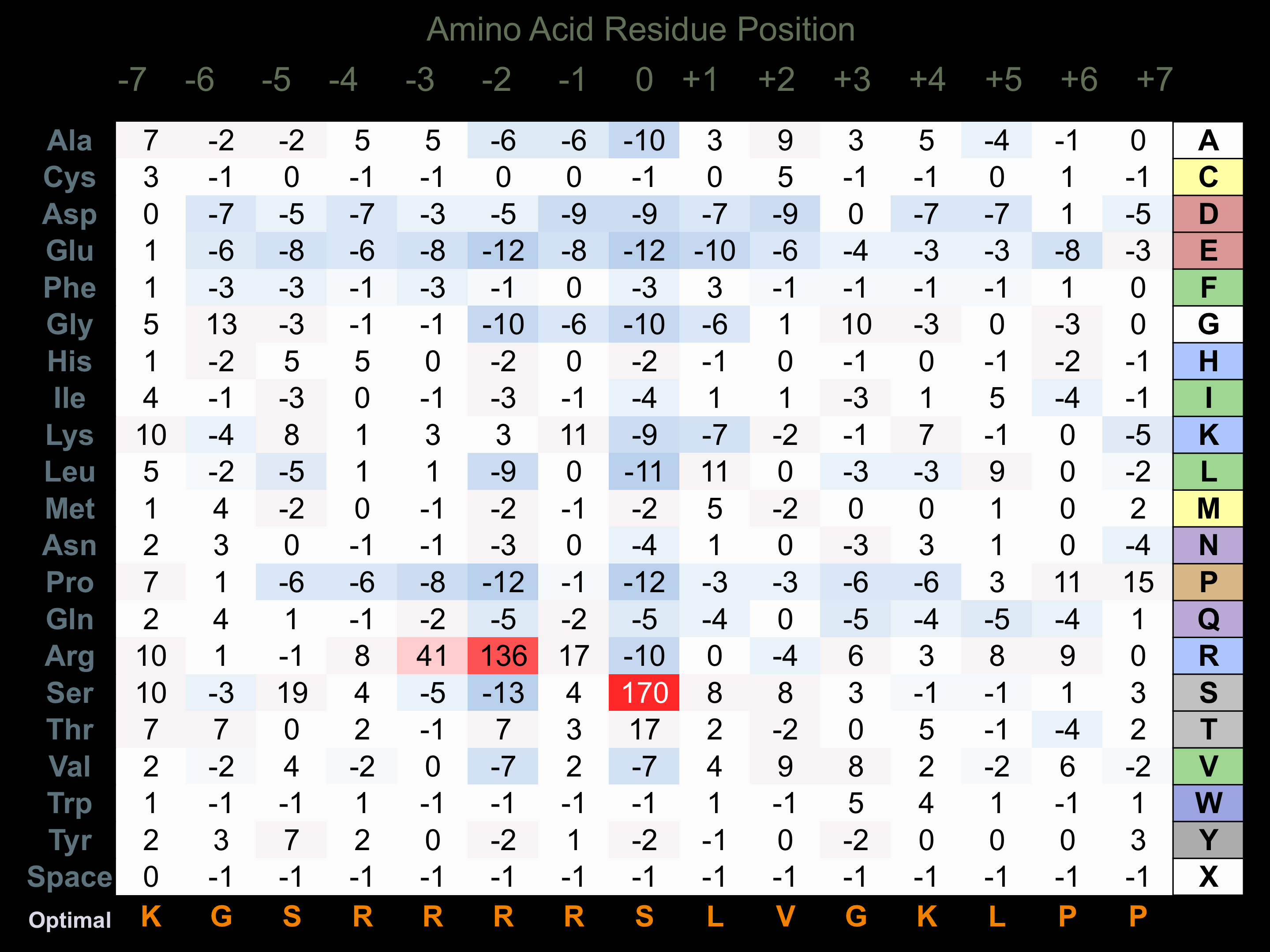
Matrix Type:
Experimentally derived from alignment of 87 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Giant cell glioblastomas (GC); polyploidy; neuroblastomas (NB), susceptibility
Comments:
AurB may be a tumour requiring protein (TRP). AurB is overexpressed in colorectal cancers and other tumour cell lines, which was thought to lead to multinuclearity, increased ploidy, and a predisposition to cancer. Through its dominant-negative effect on cytokinesis, disruptive regulation of its expression is potentially responsible for the perturbing the chromosomal integrity in cancer cells. K106R mutation leads to loss of phosphotransferase activity, and severely impairs mitosis. Its low rate of mutation and down-regulation in human cancers supports it identification as a tumour-requiring protein, and as a target for cancer drug development.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +60, p<0.064); Bladder carcinomas (%CFC= +93, p<0.028); Breast sporadic basal-like cancer (BLC) (%CFC= +52, p<0.0005); Cervical cancer (%CFC= -52, p<0.0001); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -76, p<0.0002); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +48, p<0.0002); Lung adenocarcinomas (%CFC= +137, p<0.0008); Prostate cancer - metastatic (%CFC= +73, p<0.0001); Prostate cancer - primary (%CFC= +305, p<0.0002); Skin squamous cell carcinomas (%CFC= +117, p<0.027); and Vulvar intraepithelial neoplasia (%CFC= +161, p<0.0005). The COSMIC website notes an up-regulated expression score for AurB in diverse human cancers of 861, which is 1.9-fold of the average score of 462 for the human protein kinases. In particular, in 266 ovarian cancer specimens analyzed 86% showed AurB over-expression. The down-regulated expression score of 3 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25543 diverse cancer specimens. This rate is only -9 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.27 % in 1093 large intestine cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R284H (4)
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.