Nomenclature
Short Name:
BMPR1B
Full Name:
Bone morphogenetic protein receptor type IB
Alias:
- BMR1B
- EC 2.7.11.30
- Kinase ALK6
- ACVR1
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
STKR
SubFamily:
Type1
Structure
Mol. Mass (Da):
56,930
# Amino Acids:
502
# mRNA Isoforms:
2
mRNA Isoforms:
60,582 Da (532 AA; O00238-2); 56930 Da (502 AA; O00238)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
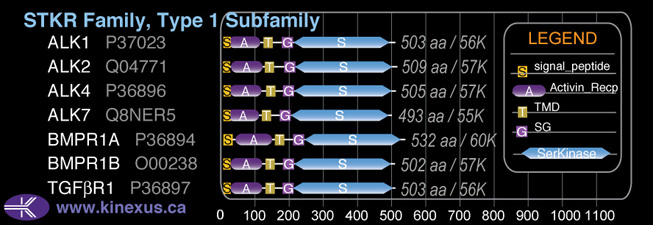
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 22 | signal_peptide |
| 17 | 110 | Activin_recp |
| 126 | 148 | TMD |
| 174 | 203 | GS |
| 204 | 493 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
T109.
Serine phosphorylated:
S300, S402.
Threonine phosphorylated:
T97.
Tyrosine phosphorylated:
Y290.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 94
94
1649
28
1953
 2
2
37
11
50
 5
5
82
6
139
 13
13
230
112
355
 29
29
503
33
427
 0.5
0.5
9
55
10
 7
7
120
39
337
 40
40
701
36
2073
 11
11
198
10
198
 7
7
122
99
155
 2
2
29
29
50
 26
26
450
132
557
 2
2
30
22
77
 0.9
0.9
15
7
12
 2
2
35
26
53
 1
1
19
18
14
 12
12
202
256
307
 2
2
42
18
113
 3
3
58
89
58
 20
20
355
112
318
 6
6
113
25
160
 2
2
27
27
60
 4
4
62
20
81
 3
3
45
18
78
 4
4
65
25
108
 100
100
1750
72
4145
 1
1
17
25
33
 5
5
81
16
116
 2
2
38
17
67
 7
7
121
42
120
 20
20
346
18
249
 18
18
319
31
472
 7
7
125
75
333
 38
38
665
83
623
 6
6
109
48
129
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99
99
99.4
99 99.2
99.2
99.8
99 -
-
-
99 -
-
-
98 71.6
71.6
83.5
98 -
-
-
- 98.2
98.2
99.6
98 69
69
80.5
98 -
-
-
- 68.2
68.2
80.5
- 91.4
91.4
95
91 33.5
33.5
52.1
89.5 78.3
78.3
85.3
81 -
-
-
- -
-
-
53 -
-
-
- 31.6
31.6
49.2
41 51.2
51.2
68.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BMP4 - P12644 |
| 2 | BMP2 - P12643 |
| 3 | BMP7 - P18075 |
| 4 | GDF5 - P43026 |
| 5 | BMPR2 - Q13873 |
| 6 | TGFBR1 - P36897 |
| 7 | BMP15 - O95972 |
| 8 | BMP6 - P22004 |
| 9 | BMPR1A - P36894 |
| 10 | GDF6 - Q6KF10 |
| 11 | TRAF6 - Q9Y4K3 |
| 12 | STAMBP - O95630 |
| 13 | GDF9 - O60383 |
| 14 | AMHR2 - Q16671 |
| 15 | SNX6 - Q9UNH7 |
Regulation
Activation:
Activated by binding bone morphogenetic protein (BMP), which induces dimerization and autophosphorylation. Binding of BMP/OP-1 to BMPR1B (a Type II receptor) induces activation of its kinase activity to phosphorylate and activate the kinase catalytic activity of the associated type I receptor.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
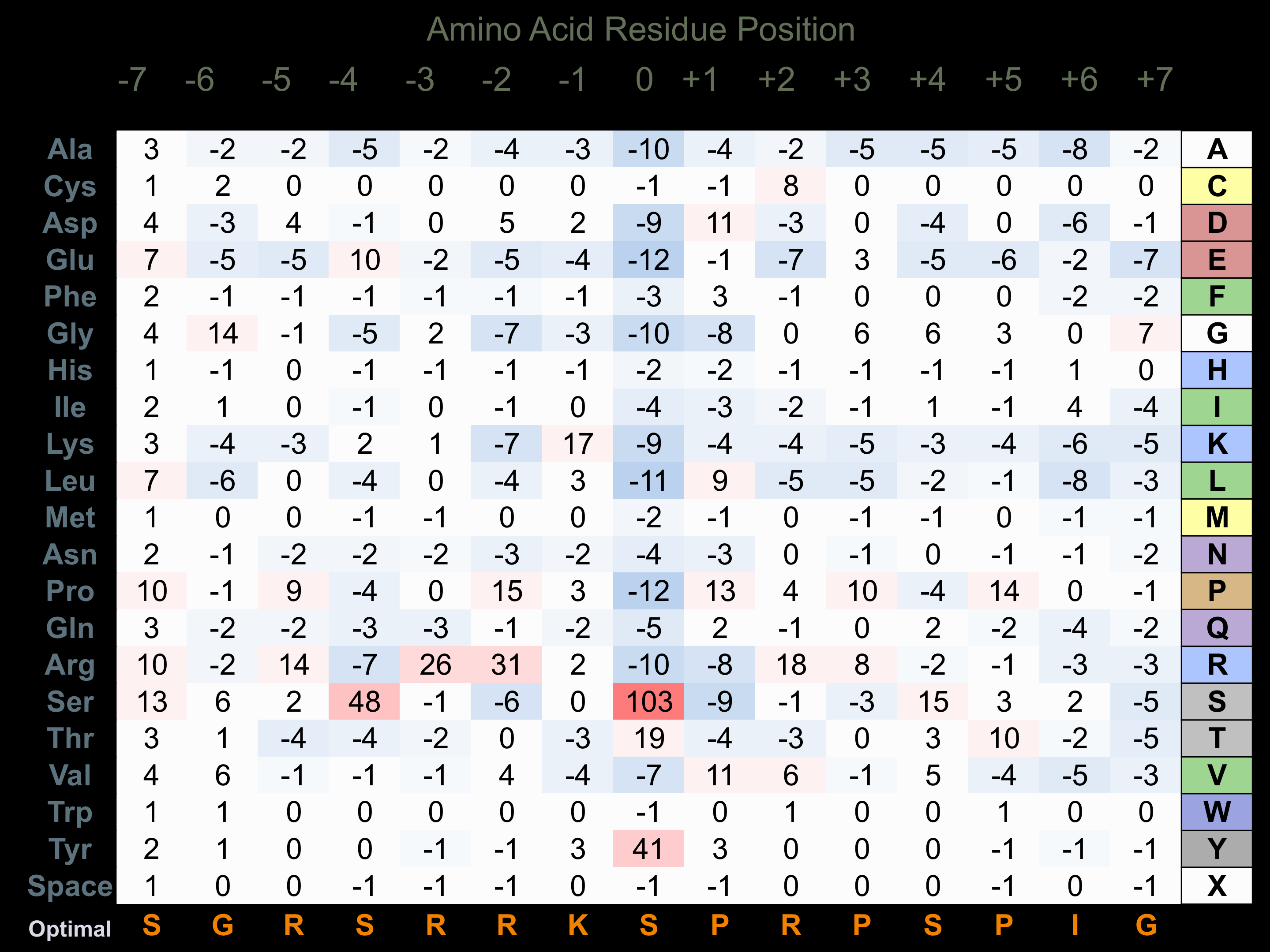
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, developmental disorders
Specific Diseases (Non-cancerous):
Type A2 brachydactyly (BDA2)
Comments:
Type A2 brachydactyly (BDA2) is an autosomal dominant hand malformation characterized by shortening and lateral deviation of the index fingers and possible shortening and deviation of the first and second toes. I200K (kinase-deficient), R486W (normal kinase activity) and R486Q mutations of BMPR1B are linked with BDA2.
Specific Cancer Types:
Prostate
Comments:
BMPR1B protein levels are down-regulated in tested cancerous prostate tissues from patients given endocrine therapy.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for BMPR1B in diverse human cancers of 374, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25346 diverse cancer specimens. This rate is very similar (+ 9% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 864 skin cancers tested; 0.39 % in 1270 large intestine cancers tested; 0.24 % in 589 stomach cancers tested; 0.17 % in 603 endometrium cancers tested; 0.15 % in 1956 lung cancers tested; 0.14 % in 1316 breast cancers tested; 0.11 % in 1467 pancreas cancers tested; 0.07 % in 1512 liver cancers tested.
Frequency of Mutated Sites:
None > 4 in 21,852 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

