Nomenclature
Short Name:
BMPR2
Full Name:
Bone morphogenetic protein receptor type II
Alias:
- BMP type II receptor
- BMPR3
- BRK-3
- EC 2.7.11.30
- PPH1
- T-ALK
- BMPR-II
- BMR2
- Bone morphogenetic protein receptor type II precursor
- Bone morphogenetic protein receptor, type II
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
STKR
SubFamily:
Type2
Specific Links
Structure
Mol. Mass (Da):
115,201
# Amino Acids:
1038
# mRNA Isoforms:
2
mRNA Isoforms:
115,201 Da (1038 AA; Q13873); 59,963 Da (530 AA; Q13873-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
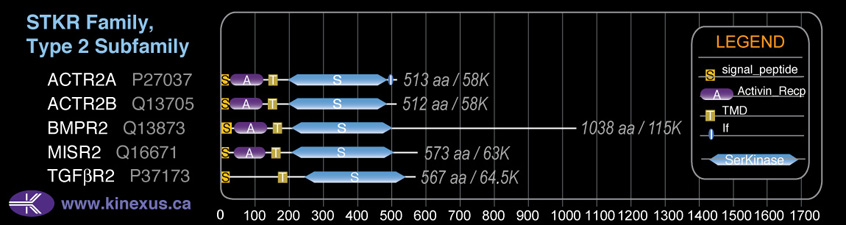
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 26 | signal_peptide |
| 35 | 131 | Activin_recp |
| 152 | 174 | TMD |
| 203 | 506 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N55, N110, N126.
Serine phosphorylated:
S375+, S513, S515, S547, S550, S574, S586, S680, S681, S757, S862, S863.
Threonine phosphorylated:
T379-, T575, T803, T805, T977.
Tyrosine phosphorylated:
Y247, Y314, Y546, Y551, Y589, Y708, Y825, Y979.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1318
53
990
 8
8
102
17
100
 26
26
349
12
327
 25
25
328
197
468
 66
66
876
58
599
 8
8
105
118
222
 9
9
115
71
219
 63
63
836
54
1789
 27
27
354
17
259
 10
10
136
161
145
 5
5
71
37
80
 63
63
830
184
692
 6
6
80
34
108
 7
7
87
11
87
 8
8
102
31
118
 8
8
109
34
119
 7
7
94
354
178
 11
11
149
19
146
 8
8
109
141
115
 53
53
698
216
609
 10
10
136
30
193
 11
11
140
34
201
 10
10
129
22
142
 13
13
174
21
226
 7
7
94
29
130
 54
54
711
128
976
 9
9
113
37
179
 9
9
122
19
97
 19
19
248
20
290
 7
7
91
70
78
 51
51
672
24
660
 29
29
381
50
451
 7
7
94
118
333
 64
64
847
161
704
 4
4
56
87
45
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.9
99.9
99.9
100 99.1
99.1
99.3
99 -
-
-
96 -
-
-
95 71.6
71.6
72
96.5 -
-
-
- 96.6
96.6
98.2
97 -
-
-
96 -
-
-
- -
-
-
- -
-
-
90 -
-
-
80 67.3
67.3
76.5
72 -
-
-
- 29.8
29.8
46.4
35 27.9
27.9
46.4
- -
-
-
- 29.1
29.1
46.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BMP4 - P12644 |
| 2 | BMP6 - P22004 |
| 3 | BMPR1B - O00238 |
| 4 | RABGGTA - Q92696 |
| 5 | TOPBP1 - Q92547 |
| 6 | CCDC89 - Q8N998 |
| 7 | GDF6 - Q6KF10 |
| 8 | KLK6 - Q92876 |
| 9 | GDF9 - O60383 |
| 10 | LSP1 - P33241 |
| 11 | MERTK - Q12866 |
| 12 | NBEA - Q8NFP9 |
| 13 | PDZRN3 - Q9UPQ7 |
| 14 | ACVR1 - Q04771 |
| 15 | ACVR1B - P36896 |
Regulation
Activation:
Activated by binding bone morphogenetic protein. This type II receptor phosphorylates and activates type I receptors which autophosphorylate, then phosphorylate SMAD transcriptional regulators.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
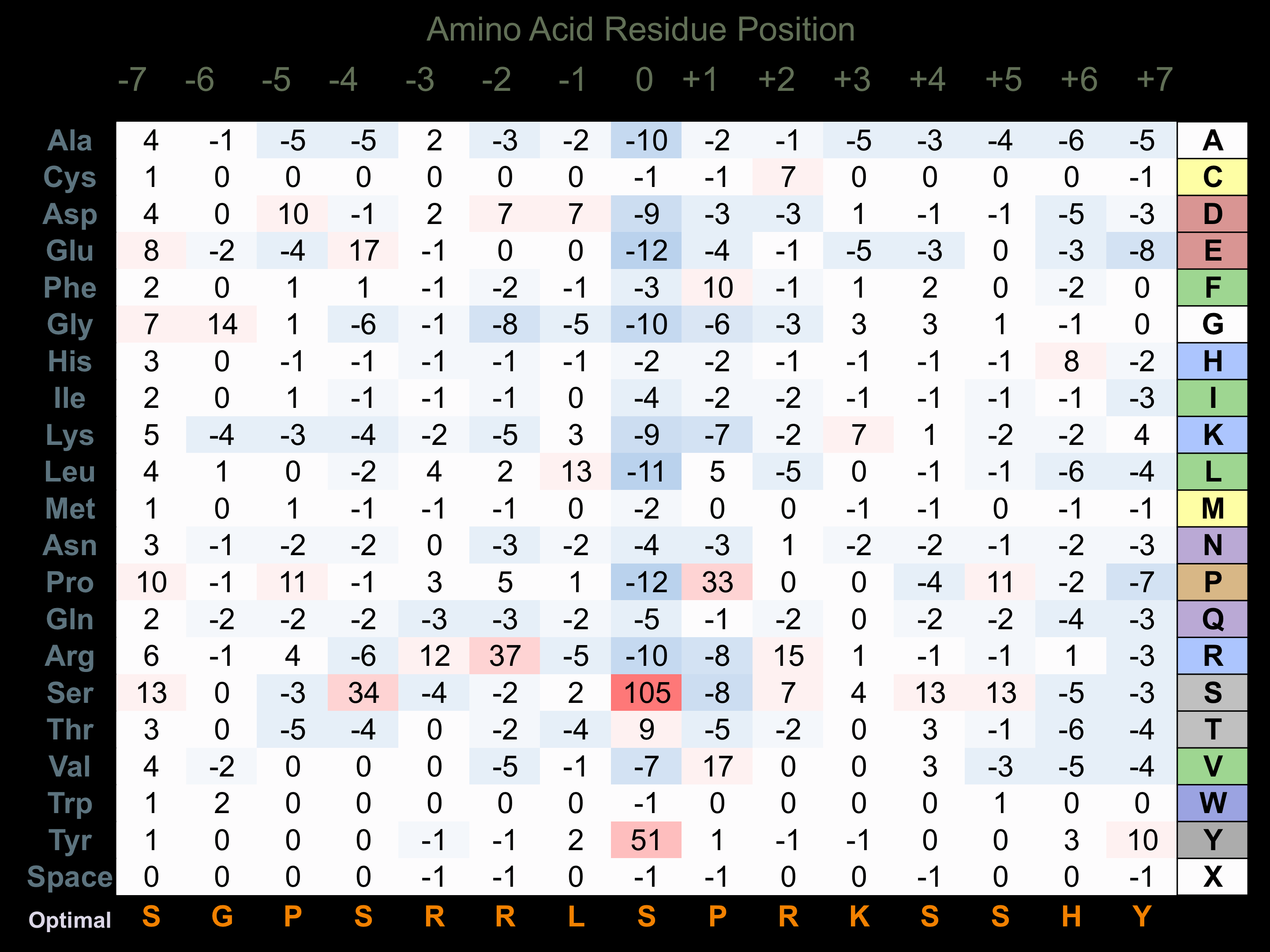
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Nintedanib | Kd = 56 nM | 9809715 | 502835 | 22037378 |
| PP242 | Kd = 89 nM | 25243800 | 22037378 | |
| Lestaurtinib | Kd = 200 nM | 126565 | 18183025 | |
| SU14813 | Kd = 200 nM | 10138259 | 1721885 | 18183025 |
| JNJ-28312141 | Kd = 310 nM | 22037378 | ||
| KW2449 | Kd = 370 nM | 11427553 | 1908397 | 22037378 |
| Sunitinib | Kd = 570 nM | 5329102 | 535 | 18183025 |
| Bosutinib | Kd = 580 nM | 5328940 | 22037378 | |
| JNJ-7706621 | Kd = 880 nM | 5330790 | 191003 | 18183025 |
| Ruxolitinib | Kd = 930 nM | 25126798 | 1789941 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Staurosporine | Kd = 2.3 µM | 5279 | 22037378 | |
| TG101348 | Kd = 2.9 µM | 16722836 | 1287853 | 22037378 |
| Dovitinib | Kd = 3.1 µM | 57336746 | 18183025 | |
| NVP-TAE684 | Kd = 3.9 µM | 16038120 | 509032 | 22037378 |
Disease Linkage
General Disease Association:
Cardiovascular disorders
Specific Diseases (Non-cancerous):
Pulmonary hypertension, familial primary, 1, with or without Hht; Pulmonary hypertension, primary, fenfluramine or dexfenfluramine-associated; Primary pulmonary hypertension; Hypertension; Pulmonary venoocclusive disease; Heritable pulmonary arterial hypertension; Hereditary hemorrhagic telangiectasia; Mixed connective tissue disease; Connective tissue disease; Pulmonary venoocclusive disease 1; Eisenmenger syndrome
Comments:
Mutations in the BMPR2 gene have been reported in patients with pulmonary hypertension, inlcuding frameshift mutations producing truncated proteins (2579delT, 44delC), substituation mutations resulting in premature termination (R899X, S73X, R873X, R332X, R211X, Y40X, Q433X, E195X), deletion/insertion mutations, and substitution mutations (C118W, C347Y, D485G, R491W, R491Q, C123R, C123S, R899P, G182D, N202Y). The various mutations are associated with a loss-of-function in the protein, and reduced BMP signalling. Interestingly, smooth muscle contractility can be stimulated by TGF-beta and BMP signalling, providing a potential mechanistic link. Furthermore, activation of the BMPR2 protein requires BMPR1A, another transmembrane receptor of the BMP receptor family. BMPR1A expression is significantly reduced in lung tissue from patients with pulmonary hypertension, indicating that the disease is linked to defects in signalling pathways involving BMPR2 and BMPR1A. In animal studies, mice with experimental reduced BMPR2 expression displayed increased inflammatory cell recruitment to the affected tissues as well as enhanced symptoms of pulmonary arterial hypertension. This indicates a role for BMPR2 expression in the regulation of inflammatory responses as well as a potential contribution of aberrant inflammation to the pathogenesis of pulmonary hypertension. Pulmonary hypertension is a cardiovascular disease characterized by high blood pressure in the vessels of the pulmonary system (i.e. lungs). Symptoms of this disease include shortness of breath, dizziness, fainting, and swelling in the extremities. Associated with pulmonary hypertension is an increased risk of pulmonary edema and pleural effusions.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +59, p<0.0009); Brain glioblastomas (%CFC= +481, p<0.024); Cervical cancer stage 2B (%CFC= -56, p<0.034); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +417, p<0.004); Oral squamous cell carcinomas (OSCC) (%CFC= +69, p<0.038); and Prostate cancer - metastatic (%CFC= -67, p<0.0001). The COSMIC website notes an up-regulated expression score for BMPR2 in diverse human cancers of 328, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 50 for this protein kinase in human cancers was 0.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25392 diverse cancer specimens. This rate is only 11 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.11 % in 1270 large intestine cancers tested; 0.08 % in 864 skin cancers tested; 0.04 % in 1512 liver cancers tested; 0.03 % in 603 endometrium cancers tested; 0.02 % in 1957 lung cancers tested; 0.02 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R179H (3); R179C (3).
Comments:
Over 50 deletions at the N583 site and one insertions at the R584 site are noted at the COSMIC website.

