Nomenclature
Short Name:
CCK4
Full Name:
Tyrosine-protein kinase-like 7
Alias:
- Colon carcinoma kinase-4
- CCK-4
- PTK7
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
CCK4
SubFamily:
NA
Structure
Mol. Mass (Da):
118392
# Amino Acids:
1070
# mRNA Isoforms:
6
mRNA Isoforms:
119,197 Da (1078 AA; Q13308-6); 118,392 Da (1070 AA; Q13308); 113,805 Da (1030 AA; Q13308-2); 112,261 Da (1014 AA; Q13308-4); 103,581 Da (940 AA; Q13308-3); 89,764 Da (816 AA; Q13308-5)
4D Structure:
NA
1D Structure:
Subfamily Alignment
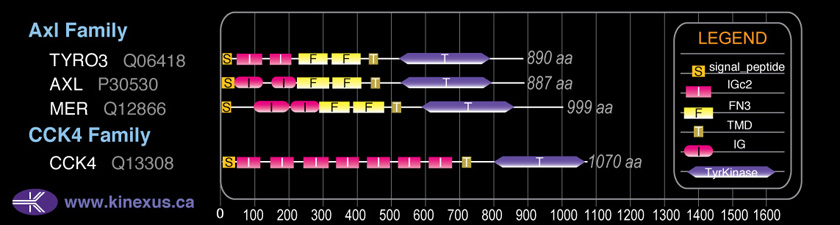
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N116, N175, N184, N214, N268, N283, N405, N463, N567, N646.
Serine phosphorylated:
S39, S40, S285, S542, S543, S784, S786.
Threonine phosphorylated:
T279, T561, T785.
Tyrosine phosphorylated:
Y872, Y877, Y960+.
Ubiquitinated:
K132, K181, K636, K804, K835, K898, K1069.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 42
42
896
28
1333
 2
2
37
10
32
 5
5
103
20
192
 67
67
1419
93
3424
 34
34
723
21
677
 6
6
133
53
213
 9
9
184
29
427
 100
100
2116
46
4262
 13
13
266
10
234
 5
5
102
61
126
 3
3
67
32
138
 26
26
550
130
593
 4
4
85
31
153
 10
10
207
9
123
 2
2
39
9
19
 1
1
20
13
16
 3
3
67
122
120
 13
13
281
26
1050
 1
1
22
62
22
 27
27
566
79
565
 3
3
63
29
100
 5
5
106
31
124
 3
3
74
29
101
 2
2
50
27
49
 7
7
140
29
137
 37
37
781
71
1453
 3
3
57
34
84
 7
7
144
27
207
 10
10
215
27
133
 7
7
146
14
79
 15
15
312
18
213
 19
19
402
26
539
 11
11
233
66
447
 29
29
610
57
593
 22
22
464
35
748
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 22.3
22.3
38.7
99 86.6
86.6
87.5
97 -
-
-
93.5 -
-
-
- 94.1
94.1
96.9
93.5 -
-
-
- 91.9
91.9
94.7
93 -
-
-
- -
-
-
- 70.6
70.6
76.3
- 71.5
71.5
82.1
66 -
-
-
71 -
-
-
53 -
-
-
- 26.6
26.6
43.2
35 35.3
35.3
52.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
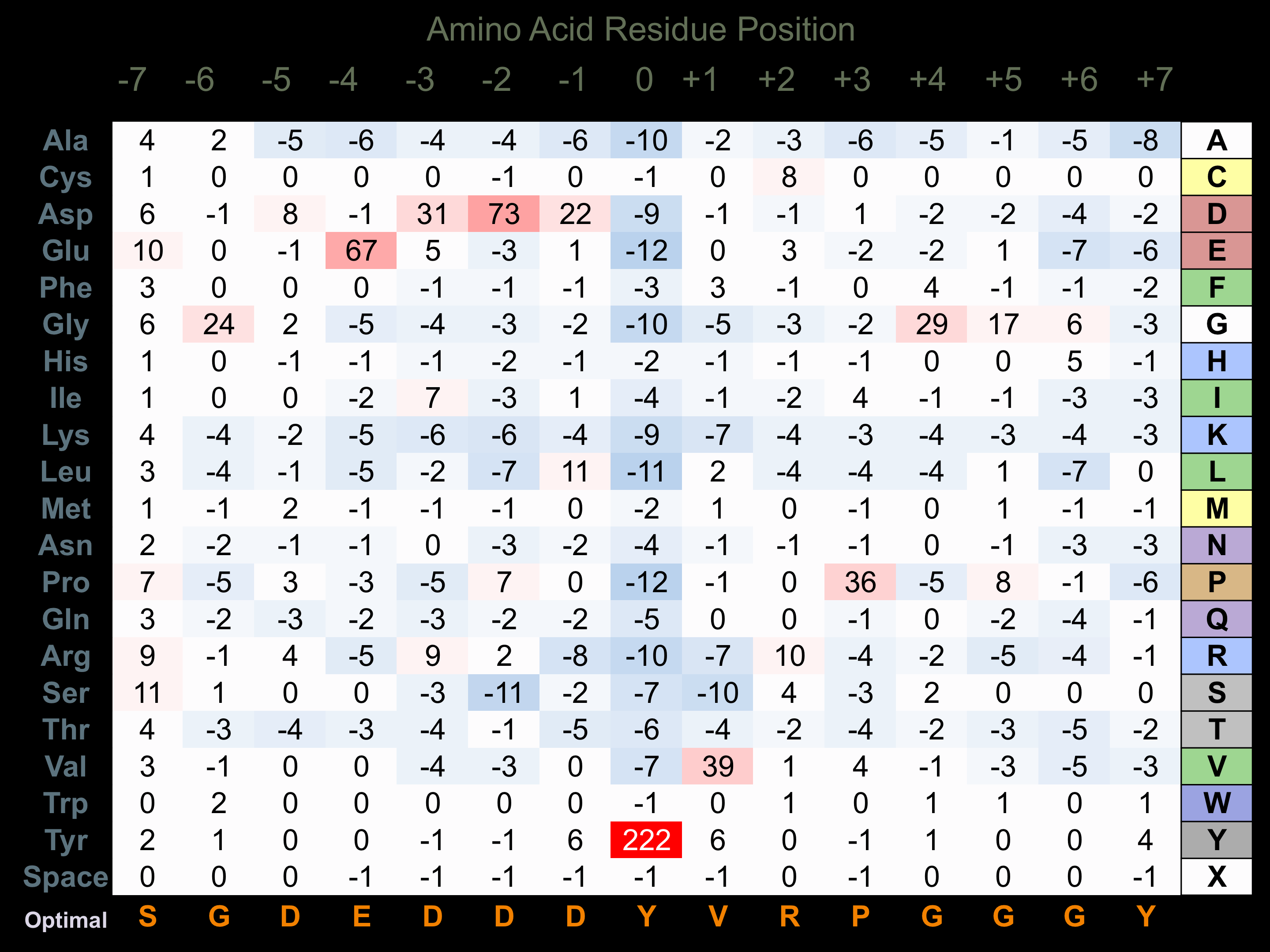
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Panic disorder; Social phobia; Agoraphobia
Comments:
Infusion of ectopic CCK4 peptide has been shown to induce panic-like symptoms in both panic disorder affected patients and non-affected controls, indicating that abnormal expression or activity of the CCK4 protein may play a role in the pathology of panic disorder and other anxiety disorders. These experimentally-induced panic symptoms can be prevented in a dose-dependent manner by the administration of selective CCK4 antagonists. In addition, a C-to-T transition mutation was observed at position -36 in the GC box of the CCK4 promoter sequence in patients with panic disorder. This suggests that the disorder may be due to aberrant expression of CCK4 rather than mutation of the protein sequence. Furthermore, ligands of the TSPO protein have been demonstrated to have potent anxiolytic activity in humans, thus they are thought to be useful for the treatment of anxiety-related disorders, including panic disorder. TSPO is an 18 KDa protein located in the outer mitochondrial membrane that mediates mitochondrial cholesterol import and neurosteroid biosynthesis. These endogenous metabolites have been shown to be significantly reduced during panic attacks and other anxiety-related events. Stimulants of this protein were shown to counteract the effects of CCK4-mediated panic attacks in an experimental context, indicating a link between CCK4 activity and neurosteroid levels in anxiety disorders. In animal studies, mice with mutated CCK4 protein display disrupted neural tube closure and abnormal stereociliary bundle orientation, indicating a role for the protein in neurodevelopment. Additionally, CCK4 protein is dynamically localized during the establishment of hair-cell polarity and appears to be a critical vertebrate regulator of planar cell polarity during development. Therefore, abnormal expression of CCK4 appears to be a contributing factor to the development of anxiety disorders (e.g. panic disorder), potentially through an interaction with neurosteroids or efffect on neurodevelopment. Panic disorder is a form of anxiety disorder characterized by panic attacks, which are sudden onset feelings of terror in non-dangerous situations. Social phobia is a phobic disorder characterized by a fear of specific public or social situations, interations with other people, or being evaluated/scrutinized. Agoraphobia is a phobic disorder characterized by the specific fear of being in a place/situation from which escape is difficult or embarrassing.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for CCK4 in diverse human cancers of 606, which is 1.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 20 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25658 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 854 skin cancers tested; 0.32 % in 1243 large intestine cancers tested; 0.17 % in 589 stomach cancers tested; 0.12 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,941 cancer specimens
Comments:
Only 3 deletions, 4 insertions or 1 complex mutation are noted on the COSMIC website.

