Nomenclature
Short Name:
CDK10
Full Name:
Cell division protein kinase 10
Alias:
- EC 2.7.11.22
- PISSLRE
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDK10
Specific Links
Structure
Mol. Mass (Da):
41,038
# Amino Acids:
360
# mRNA Isoforms:
7
mRNA Isoforms:
41,038 Da (360 AA; Q15131); 32,731 Da (289 AA; Q15131-2); 32,662 Da (289 AA; Q15131-7); 31,944 Da (283 AA; Q15131-3); 30,702 Da (272 AA; Q15131-4); 21,512 Da (188 AA; Q15131-5); 17,446 Da (154 AA; Q15131-6)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 39 | 323 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
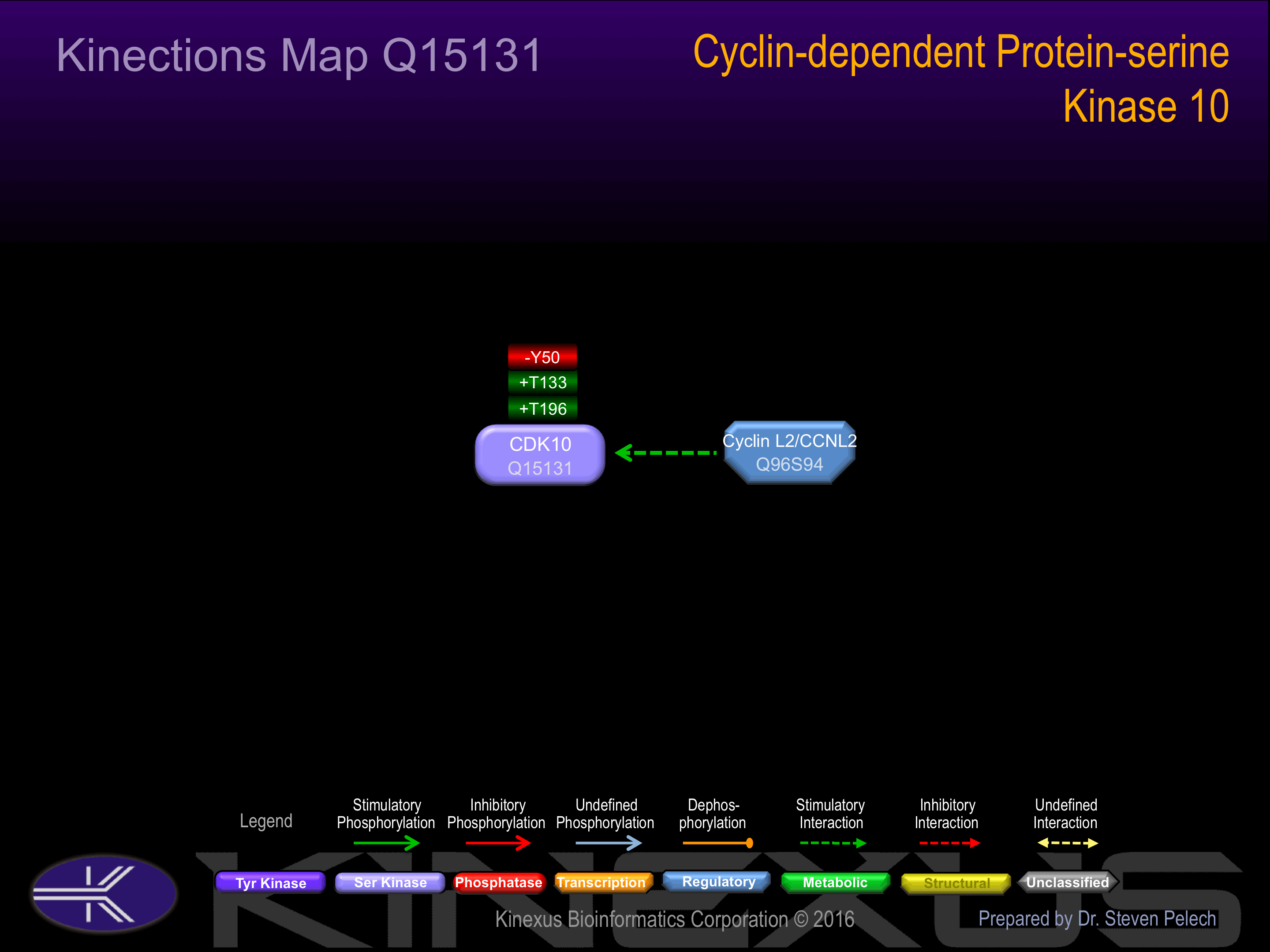
Threonine phosphorylated:
T49, T133+, T196+.
Tyrosine phosphorylated:
Y50-, Y54.
Ubiquitinated:
K68, K76.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 96
96
813
42
966
 7
7
56
20
45
 6
6
48
3
29
 43
43
365
114
501
 100
100
847
36
702
 4
4
32
111
19
 40
40
336
47
626
 79
79
667
39
601
 63
63
533
24
378
 20
20
172
112
261
 15
15
127
30
118
 87
87
741
221
644
 5
5
41
36
32
 12
12
104
15
156
 16
16
135
21
136
 10
10
85
22
49
 15
15
123
202
109
 8
8
64
14
48
 6
6
50
111
56
 61
61
516
162
546
 17
17
140
18
128
 17
17
148
24
239
 8
8
67
14
47
 7
7
58
14
47
 18
18
154
18
214
 93
93
785
62
682
 5
5
42
39
36
 8
8
71
14
66
 12
12
105
14
132
 14
14
115
42
101
 65
65
550
30
613
 79
79
668
51
604
 0.7
0.7
6
51
3
 86
86
726
83
658
 19
19
165
57
273
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
91 -
-
-
99 -
-
-
95 -
-
-
95 93
93
96.9
95 -
-
-
- 96.9
96.9
98.8
97 90.5
90.5
93.6
97 -
-
-
- -
-
-
- 82.9
82.9
89.7
85 36.9
36.9
53.6
87 60.8
60.8
69.1
80 -
-
-
- 21.3
21.3
27.8
66.5 63.9
63.9
73.6
- 25.8
25.8
36.4
- 63.7
63.7
75.8
- -
-
-
- -
-
-
- -
-
-
- 31.4
31.4
45.5
- -
-
-
- 29.7
29.7
43.6
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | RAF1 - P04049 |
| 2 | MNAT1 - P51948 |
| 3 | CCNB2 - O95067 |
| 4 | CCNC - P24863 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
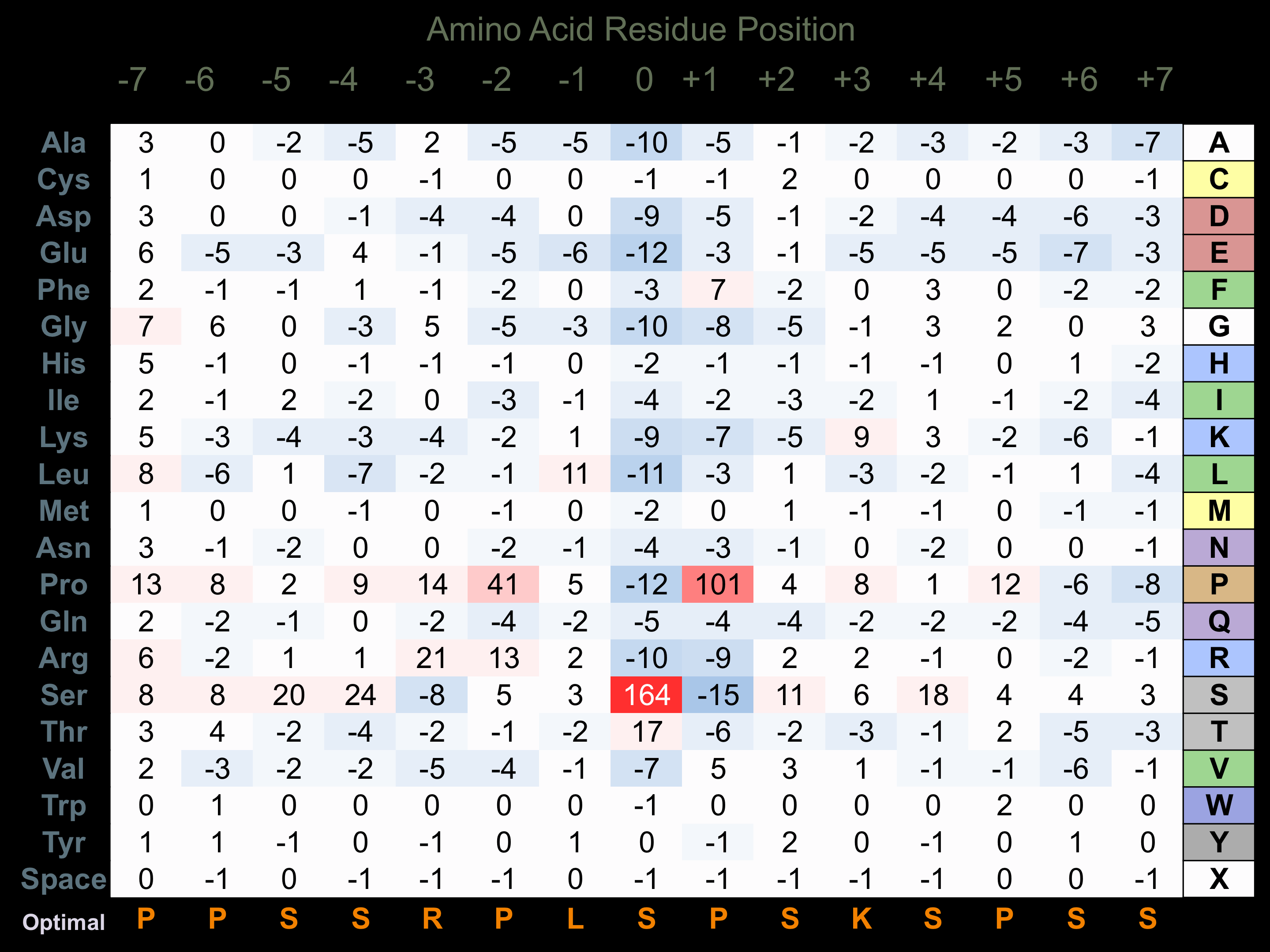
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Renal and anogenital malformations
Specific Diseases (Non-cancerous):
Star syndrome
Comments:
Star syndrome is a rare disorder which includes anomalies of the fallopian tubes, uterine lining, and uterus, as well as retinal and chorioretinal dysplasia or dystrophy, and macular dystrophy or absence or hypoplasia of the macula.
Comments:
CDK10 appears to be a tumour requiring protein (TRP). It displays one of the lowest rates of mutation in human tumours.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +56, p<0.024); Breast epithelial cell carcinomas (%CFC= +77, p<0.004); Cervical cancer (%CFC= -45, p<(0.0003); Large B-cell lymphomas (%CFC= +52, p<0.017); and Ovary adenocarcinomas (%CFC= +94, p<0.072). The COSMIC website notes an up-regulated expression score for CDK10 in diverse human cancers of 386, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 10 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0 % in 24900 diverse cancer specimens. This rate is -99 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a very low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Frequency of Mutated Sites:
Only 1 mutation: N157S.
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.