Nomenclature
Short Name:
CDK11
Full Name:
Cell division protein kinase 11
Alias:
- CDK19
- CD2L6
- Cell division cycle 2-like 6
- Cyclin-dependent kinase (CDC2-like) 11
- Death-preventing kinase
- KIAA1028; EC 2.7.11.22;;
- CDC2L6
- CDK8-like
- CDK8-like cyclin-dependent kinase
- CDK8-like cyclin-dependent kinase 11
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDK8
Structure
Mol. Mass (Da):
56,802
# Amino Acids:
502
# mRNA Isoforms:
2
mRNA Isoforms:
56,802 Da (502 AA; Q9BWU1); 49,870 Da (442 AA; Q9BWU1-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 21 | 335 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S449.
Threonine phosphorylated:
T31.
Tyrosine phosphorylated:
Y276.
Ubiquitinated:
K52.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 97
97
899
48
994
 4
4
35
21
36
 11
11
106
3
91
 53
53
496
146
643
 72
72
674
46
613
 7
7
63
120
54
 21
21
193
53
403
 73
73
681
42
864
 59
59
545
24
467
 6
6
60
129
59
 4
4
41
31
40
 84
84
785
221
671
 6
6
59
36
49
 6
6
52
15
52
 6
6
55
21
57
 5
5
45
27
49
 18
18
170
203
351
 3
3
31
13
35
 5
5
44
129
38
 62
62
577
190
597
 16
16
152
20
152
 7
7
69
26
71
 9
9
80
6
71
 10
10
94
14
56
 8
8
79
20
82
 88
88
820
81
1257
 7
7
68
39
61
 4
4
41
14
43
 8
8
79
14
76
 5
5
49
56
46
 76
76
704
30
799
 58
58
542
56
549
 14
14
131
102
178
 100
100
930
104
801
 47
47
438
61
339
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 79.5
79.5
81.2
98 -
-
-
- -
-
-
94 -
-
-
97 89.8
89.8
90.8
94 -
-
-
- 96.4
96.4
97.2
92 96.4
96.4
97.2
93 -
-
-
- 77.5
77.5
83.5
- 76.8
76.8
78.4
93 68.1
68.1
74.1
89 76.7
76.7
82.7
88 -
-
-
- 67.3
67.3
74.5
85 -
-
-
- 46.9
46.9
60.4
60 -
-
-
- -
-
-
- -
-
-
- -
-
-
41 37.9
37.9
52.6
44 -
-
-
- -
-
-
47.5
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
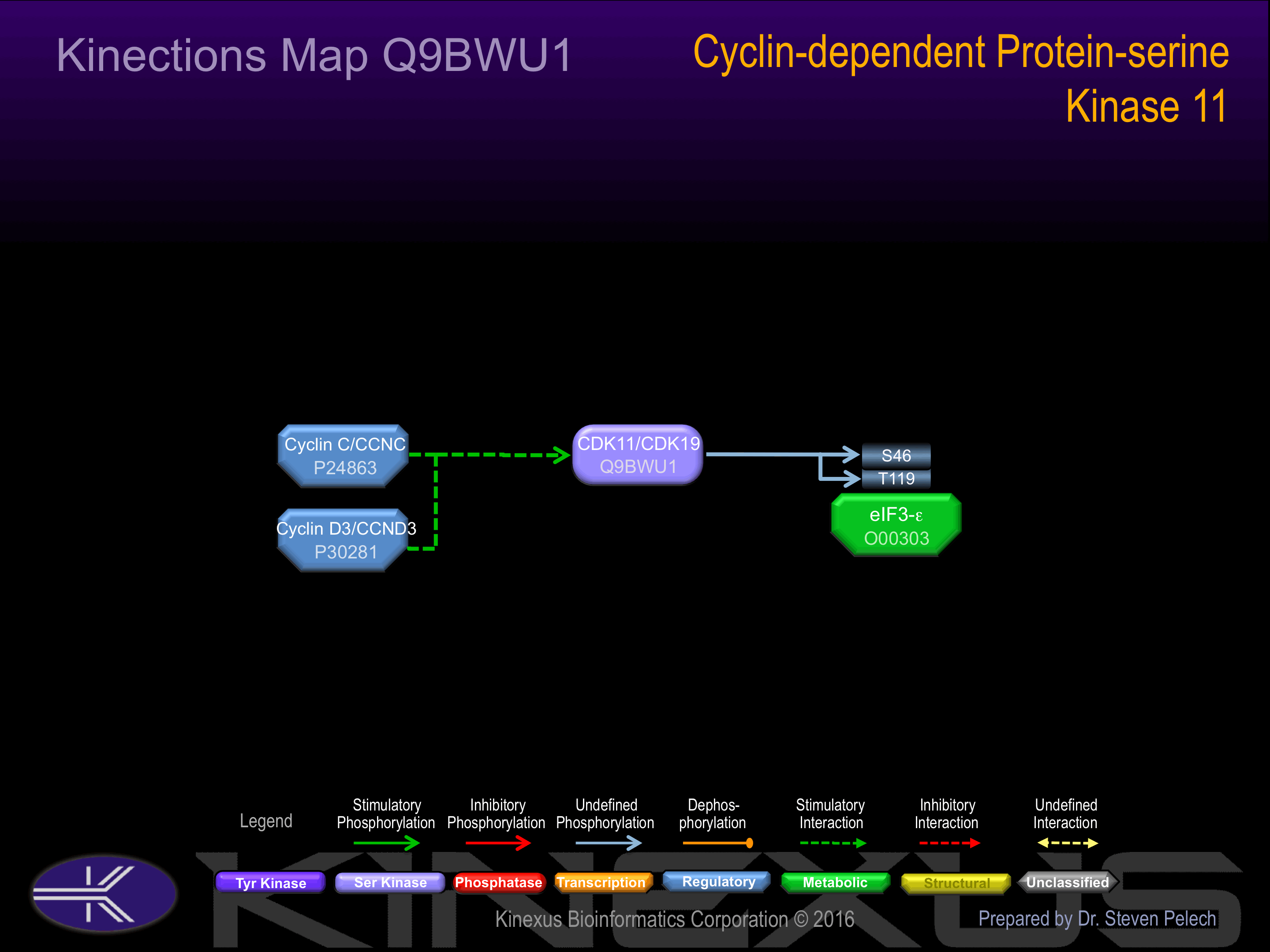
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | EIF3F - O00303 |
| 2 | SFRS7 - Q16629 |
| 3 | CDK7 - P50613 |
| 4 | RB1CC1 - Q8TDY2 |
| 5 | MYCBPAP - Q8TBZ2 |
| 6 | MED28 - Q9H204 |
| 7 | MED26 - O95402 |
| 8 | MED4 - Q9NPJ6 |
| 9 | MED29 - Q9NX70 |
| 10 | MED1 - Q15648 |
| 11 | MED15 - Q96RN5 |
| 12 | MED17 - Q9NVC6 |
| 13 | MED7 - O43513 |
| 14 | MED9 - Q9NWA0 |
| 15 | MED27 - Q6P2C8 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
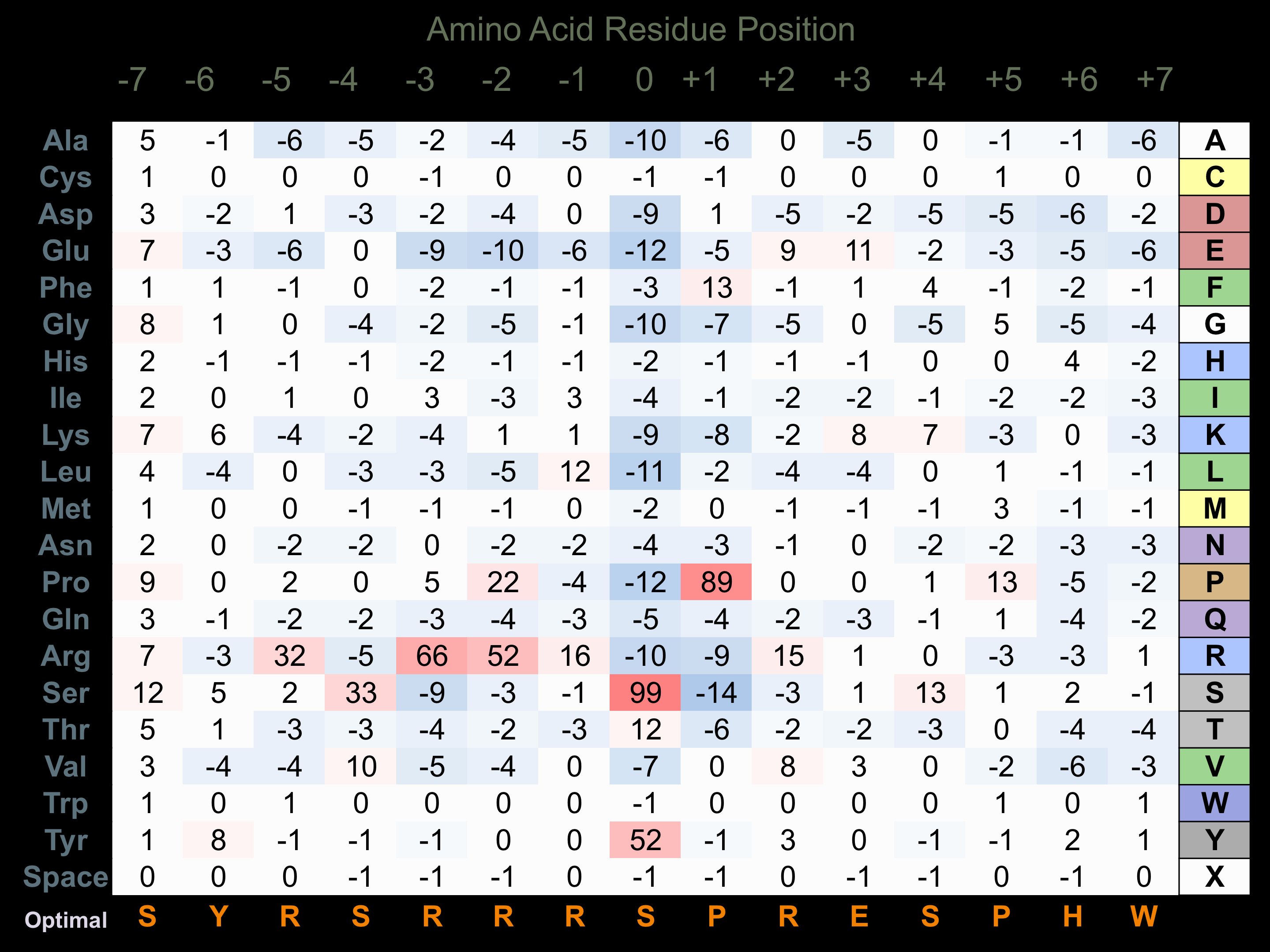
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AST-487 | Kd = 1.5 nM | 11409972 | 574738 | 18183025 |
| Alvocidib | Kd = 57 nM | 9910986 | 428690 | 18183025 |
| Linifanib | Kd = 74 nM | 11485656 | 223360 | 18183025 |
| Foretinib | Kd = 87 nM | 42642645 | 1230609 | 22037378 |
| Staurosporine | Kd = 190 nM | 5279 | 18183025 | |
| Doramapimod | Kd = 200 nM | 156422 | 103667 | 18183025 |
| Sorafenib | Kd = 250 nM | 216239 | 1336 | 18183025 |
| PLX4720 | Kd = 700 nM | 24180719 | 1230020 | 22037378 |
| AC1NS7CD | Kd = 950 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 950 nM | 3025986 | 296468 | 18183025 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| CP724714 | Kd = 1.1 µM | 9874913 | 483321 | 18183025 |
| A674563 | Kd = 1.3 µM | 11314340 | 379218 | 22037378 |
| Vatalanib | Kd = 1.5 µM | 151194 | 101253 | 22037378 |
| AT7519 | Kd = 3.3 µM | 11338033 | 22037378 |
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +202, p<0.102); Clear cell renal cell carcinomas (cRCC) (%CFC= +59, p<0.061); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -77, p<0.0001); Prostate cancer (%CFC= -55, p<0.073); Prostate cancer - metastatic (%CFC= +140, p<0.0001); Prostate cancer - primary (%CFC= -83, p<0.0001); and Skin melanomas - malignant (%CFC= +105, p<0.003). The COSMIC website notes an up-regulated expression score for CDK11 in diverse human cancers of 440, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 80 for this protein kinase in human cancers was 1.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24939 diverse cancer specimens. This rate is very similar (+ 4% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 4 in 20,222 cancer specimens
Comments:
Only 1 deletion, 1 complex mutation and no insertions mutations are noted on the COSMIC website.