Nomenclature
Short Name:
CDKL1
Full Name:
Serine-threonine-protein kinase KKIALRE
Alias:
- Cyclin-dependent kinase-like 1
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDKL
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
41671
# Amino Acids:
357
# mRNA Isoforms:
2
mRNA Isoforms:
41,671 Da (357 AA; Q00532); 41,671 Da (357 AA; Q00532-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
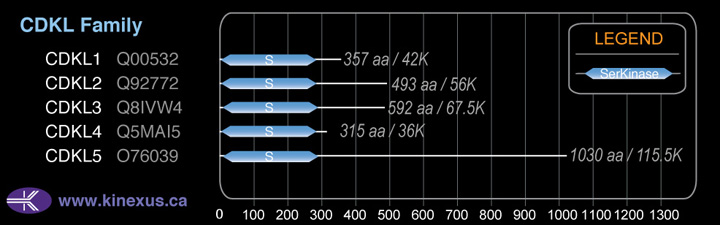
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 287 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Threonine phosphorylated:
T26.
Tyrosine phosphorylated:
Y161.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
762
28
1077
 1
1
9
14
8
 25
25
189
9
146
 16
16
122
108
279
 54
54
412
27
225
 0.7
0.7
5
55
6
 26
26
198
43
396
 78
78
594
33
1646
 36
36
275
10
281
 9
9
66
84
63
 2
2
13
25
13
 54
54
409
139
505
 6
6
46
20
67
 2
2
14
9
15
 3
3
24
21
22
 1
1
9
19
10
 3
3
24
201
19
 3
3
25
17
21
 2
2
13
75
13
 50
50
379
107
309
 5
5
38
22
43
 11
11
82
23
113
 5
5
36
18
36
 2
2
17
18
18
 4
4
28
24
21
 59
59
450
70
469
 3
3
25
23
36
 8
8
59
15
64
 45
45
341
16
412
 17
17
129
28
118
 66
66
502
18
368
 36
36
276
31
363
 0.4
0.4
3
41
1
 88
88
673
83
627
 3
3
24
48
9
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
99 97.7
97.7
99.1
- -
-
-
91.5 -
-
-
- 88.5
88.5
92.6
91 -
-
-
- 88.8
88.8
92.7
90 88.2
88.2
93
89.5 -
-
-
- 86.5
86.5
92.7
- 37.8
37.8
52.3
84 36.9
36.9
54.6
85 77
77
87.9
79 -
-
-
- 43.1
43.1
54.6
61 53.2
53.2
69.6
- -
-
-
62 59.5
59.5
73
- -
-
-
- 34.4
34.4
54.3
- -
-
-
- 33
33
54.6
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
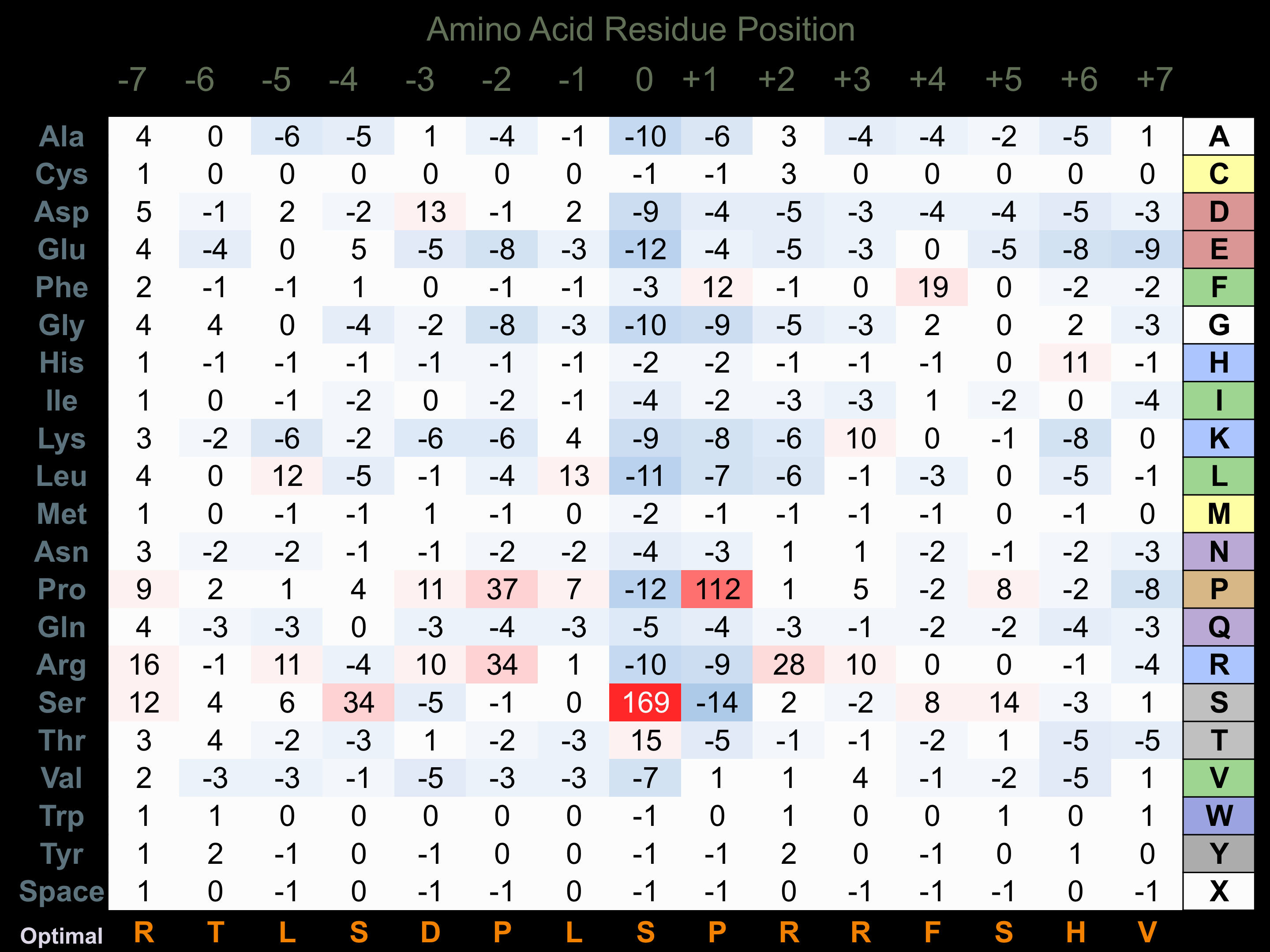
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Gastric cancer
Comments:
CDKL1 may be an oncoprotein (OP). Significantly elevated CDKL1 expression was observed in gastric cancer tissue specimens. In addition, loss of CDKL1 function in gastric cancer cell lines (SGC7901 and MGC-803) resulted in impaired cell proliferation and enhanced apoptosis, indicating that CDKL1 functions as an oncoprotein. Furthermore, siRNA-mediated knockdown of CDKL1 expression stimulated the pro-apoptotic factor Bcl-2-interacting killer (BIK) and reduced cellular expression of the proliferating cell nuclear antigen (PCNA), indicating reduced cellular proliferation. Therefore, CDKL1 is thought to play a key role in gastric cancer cell proliferation and survival.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for CDKL1 in diverse human cancers of 369, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 2 for this protein kinase in human cancers was 97% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24727 diverse cancer specimens. This rate is only -31 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.29 % in 1270 large intestine cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,012 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

