Nomenclature
Short Name:
CLIK1L
Full Name:
PDIK1L protein
Alias:
- Novel kinase
- Novel protein
- PDIK1L
- BF960819
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NKF4
SubFamily:
NA
Structure
Mol. Mass (Da):
38,546
# Amino Acids:
341
# mRNA Isoforms:
1
mRNA Isoforms:
38,546 Da (341 AA; Q8N165)
4D Structure:
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 8 | 331 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K187.
Methylated:
R16 (m1).
Serine phosphorylated:
S31.
Threonine phosphorylated:
T30.
Ubiquitinated:
K187, K271, K305, K309, K335.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 20
20
1535
9
1249
 0.1
0.1
10
4
16
 -
-
-
-
-
 5
5
377
32
1094
 8
8
627
13
587
 0.7
0.7
54
9
9
 0.7
0.7
54
13
31
 27
27
2079
8
3339
 0.03
0.03
2
3
1
 0.9
0.9
72
29
36
 0.4
0.4
30
5
39
 7
7
512
17
509
 -
-
-
-
-
 0.03
0.03
2
3
1
 0.4
0.4
30
5
37
 0.3
0.3
24
5
7
 0.7
0.7
50
68
29
 0.04
0.04
3
3
2
 0.9
0.9
67
21
32
 13
13
1000
31
675
 0.4
0.4
29
5
40
 0.4
0.4
28
5
39
 -
-
-
-
-
 0.03
0.03
2
3
0
 0.8
0.8
62
5
89
 10
10
735
22
706
 0.03
0.03
2
3
0
 0.03
0.03
2
3
1
 0.01
0.01
1
3
0
 0.4
0.4
31
14
28
 20
20
1522
12
33
 100
100
7670
11
8903
 5
5
388
39
819
 13
13
978
26
780
 0.5
0.5
36
22
42
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 59.5
59.5
71.5
100 59.5
59.5
71.5
100 -
-
-
99 -
-
-
- 51.5
51.5
62
99 -
-
-
- 99.4
99.4
100
99 -
-
-
100 -
-
-
- 77.9
77.9
81.3
- 20.5
20.5
37.6
96 82.9
82.9
92.9
- 82.6
82.6
91.7
83 -
-
-
- 20.3
20.3
39.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
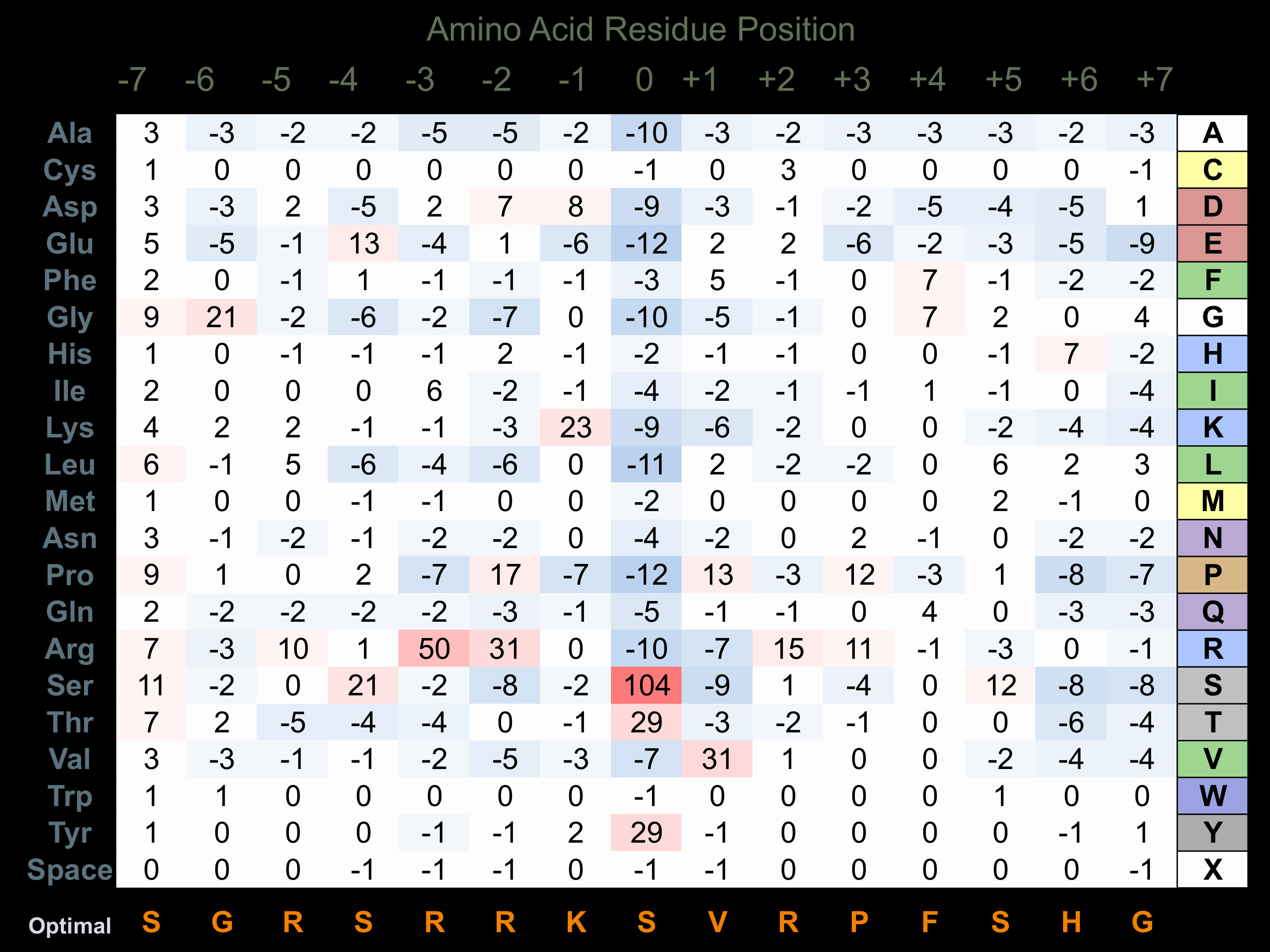
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, facial deformation, hearing and sight loss; fatty acid accumulation, and neurological disorders
Specific Diseases (Non-cancerous):
Stickler syndrome; Parkinson’s disease; Gaucher disease; Schizophrenia; Usher syndrome
Comments:
Stickler syndrome manifests with symptoms including a distinct, flat, facial appearance, hearing impairment, eye deformation, and joint issues. Gaucher Disease occurs with fatty acid accumulation in cells leading to fatigue, bruising, anemia, low platelet count, and liver or spleen enlargement. Usher syndrome leads to hearing and sight loss.
Specific Cancer Types:
Familial adenomatous polyposis (FAP)
Comments:
CLIK1L has been linked to Familial Adenomatous Polyposis (FAP), which is cancer of the large intestine, beginning with polyps in teen years, and leading to malignant tumours if left untreated.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for CLIK1L in diverse human cancers of 335, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 142 for this protein kinase in human cancers was 2.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24727 diverse cancer specimens. This rate is only -37 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.24 % in 603 endometrium cancers tested; 0.18 % in 1270 large intestine cancers tested; 0.1 % in 881 prostate cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,010 cancer specimens
Comments:
Only 2 deletions, no insertions or complex mutations are noted on the COSMIC website.

