Nomenclature
Short Name:
CaMK1b
Full Name:
PNCK pregnancy up-regulated non-ubiquitously expressed CaM kinase
Alias:
- CaM kinase I beta
- CaM kinase IB
- KCC1B
- PNCK
- Pregnancy up-regulated non-ubiquitously expressed CaM kinase
- CaMK1-beta
- CaM-KI beta
- CaMKI-beta
- EC 2.7.11.17
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMK1
SubFamily:
NA
Structure
Mol. Mass (Da):
38,500
# Amino Acids:
343
# mRNA Isoforms:
5
mRNA Isoforms:
46,957 Da (426 AA; Q6P2M8-5); 40,838 Da (366 AA; Q6P2M8-2); 40,356 Da (360 AA; Q6P2M8-6); 38,500 Da (343 AA; Q6P2M8); 27,161 Da (241 AA; Q6P2M8-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
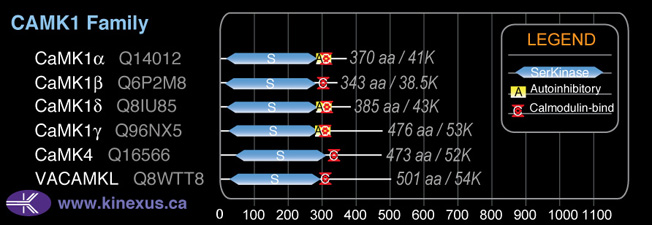
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 15 | 270 | Pkinase |
| 290 | 311 | CaM_binding |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Threonine phosphorylated:
T171+, T175-.
Tyrosine phosphorylated:
Y229.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1886
6
1816
 0.1
0.1
2
1
0
 -
-
-
-
-
 20
20
375
29
818
 45
45
845
7
69
 0.3
0.3
5
9
3
 0.3
0.3
5
10
3
 36
36
681
2
158
 -
-
-
-
-
 8
8
142
26
144
 13
13
238
2
110
 59
59
1122
8
409
 -
-
-
-
-
 -
-
-
-
-
 13
13
242
2
46
 2
2
32
5
9
 55
55
1045
65
572
 -
-
-
-
-
 3
3
56
18
45
 20
20
384
28
214
 82
82
1550
2
481
 17
17
317
2
34
 -
-
-
-
-
 -
-
-
-
-
 27
27
500
2
122
 69
69
1296
18
1803
 -
-
-
-
-
 -
-
-
-
-
 -
-
-
-
-
 5
5
96
14
35
 -
-
-
-
-
 35
35
652
11
1317
 0.1
0.1
2
12
0
 41
41
774
26
669
 2
2
30
13
10
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 61.1
61.1
77
99 88.7
88.7
90.7
92 -
-
-
93 -
-
-
- 76.7
76.7
78.7
95 -
-
-
- 95.3
95.3
96.5
95 95.3
95.3
96.5
95 -
-
-
- 75.2
75.2
85.3
- 47
47
60.2
- 40.3
40.3
58.4
79 35.5
35.5
53.2
64 37
37
54.6
- 30.2
30.2
42.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 29.4
29.4
43.7
- -
-
-
50 31
31
45.3
48 -
-
-
- -
-
-
45.5
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by Ca2+/calmodulin.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
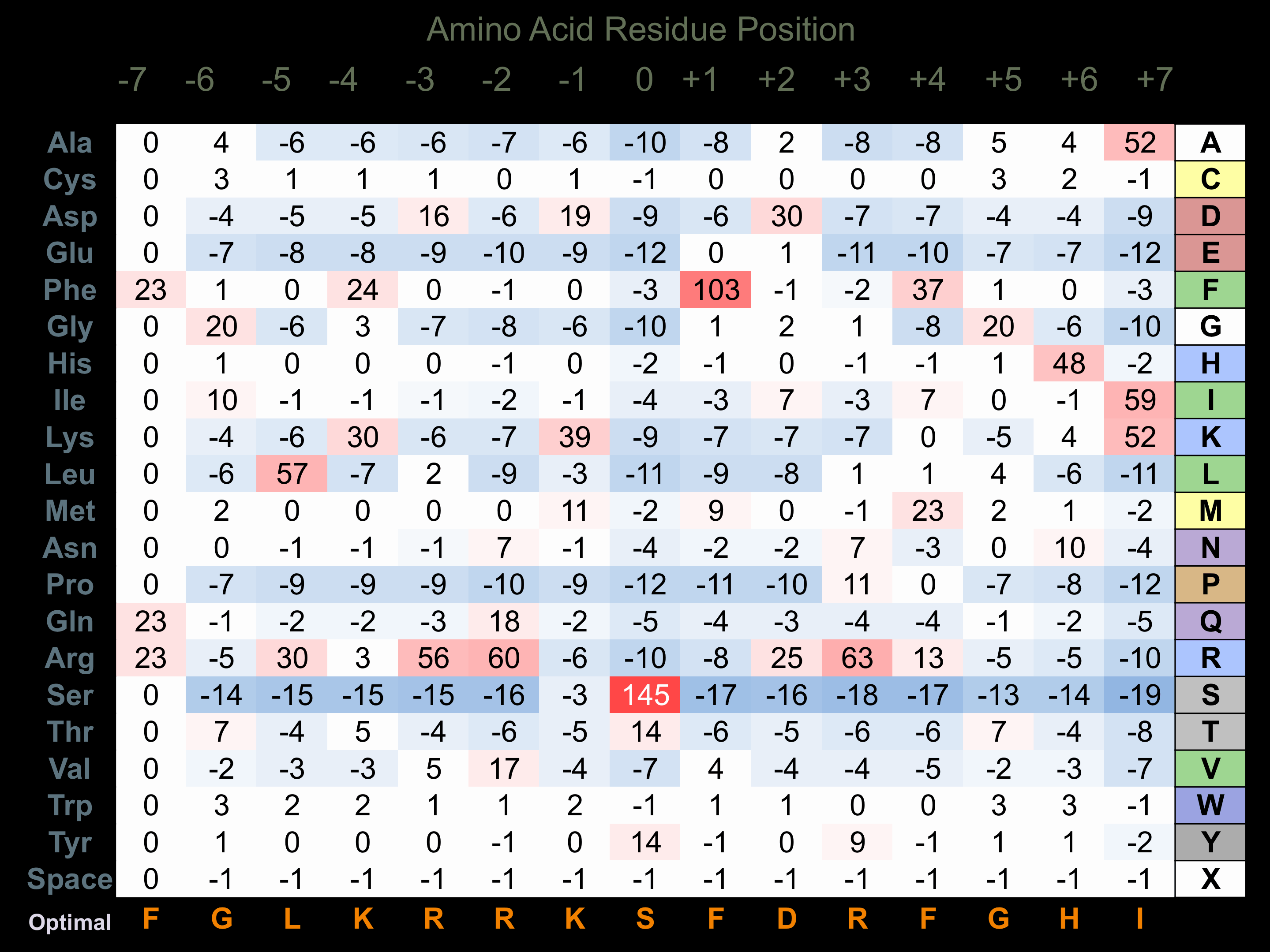
Matrix Type:
Experimentally derived from alignment of 3 known protein substrate phosphosites and 11 peptides phosphorylated by recombinant CAMK1-beta in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | IC50 > 50 nM | 5279 | 22037377 | |
| SB218078 | IC50 = 500 nM | 447446 | 289422 | 22037377 |
| K-252a; Nocardiopsis sp. | IC50 > 1 µM | 3813 | 281948 | 22037377 |
| Syk Inhibitor | IC50 > 1 µM | 6419747 | 104279 | 22037377 |
| Syk Inhibitor II | IC50 > 1 µM | 16760670 | 22037377 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 |
Disease Linkage
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for CaMK1b in diverse human cancers of 620, which is 1.3-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24644 diverse cancer specimens. This rate is only 21 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.35 % in 1259 large intestine cancers tested; 0.29 % in 603 endometrium cancers tested.
Frequency of Mutated Sites:
None > 3 in 19,927 cancer specimens
Comments:
Only 1 deletion, 1 insertion and 1 complex mutation are noted on the COSMIC website.

