Nomenclature
Short Name:
CaMK2a
Full Name:
Calcium-calmodulin-dependent protein kinase type II alpha chain
Alias:
- Calcium/calmodulin-dependent protein kinase II alpha
- Calcium/calmodulin-dependent protein kinase II alpha-B subunit
- CaMK-II alpha subunit
- CaMKII-alpha
- CaMKIINalpha
- Kinase CaMK2-alpha;CaM-kinase II alpha chain; EC 2.7.11.17; KCC2A; KIAA0968;
- Calcium/calmodulin-dependent protein kinase type II alpha chain
- CaM kinase II alpha subunit
- CaMK2-alpha
- CAMKA
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMK2
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
54088
# Amino Acids:
478
# mRNA Isoforms:
2
mRNA Isoforms:
55,320 Da (489 AA; Q9UQM7-2); 54,088 Da (478 AA; Q9UQM7)
4D Structure:
CAMK2 is composed of four different chains: alpha, beta, gamma, and delta. The different isoforms assemble into homo- or heteromultimeric holoenzymes composed of 8 to 12 subunits. Interacts with BAALC, MPDZ, SYN1, CAMK2N2 and SYNGAP1
3D Structure:
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
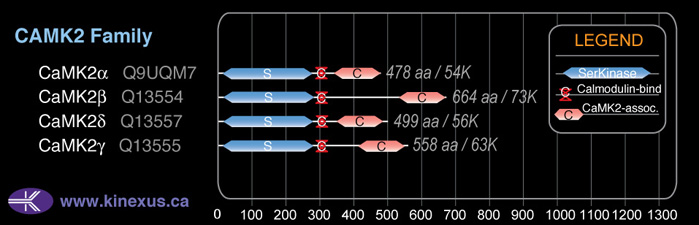
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 13 | 271 | Pkinase |
| 296 | 311 | CaM_binding |
| 346 | 473 | CaMK2-Association |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K226.
Methylated:
K258, K300.
Serine phosphorylated:
S78, S145, S234, S279, S314, S318, S331, S333, S404.
Threonine phosphorylated:
T253, T286+, T305-, T306-, T310, T334, T336, T337.
Tyrosine phosphorylated:
Y230.
Ubiquitinated:
K226.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 22
22
376
29
566
 1.4
1.4
24
13
35
 9
9
149
18
176
 51
51
869
102
973
 17
17
286
25
233
 1.1
1.1
19
78
62
 8
8
143
31
363
 34
34
581
50
793
 14
14
238
17
199
 2
2
37
70
43
 2
2
39
36
53
 35
35
593
169
619
 3
3
57
41
108
 1.4
1.4
23
9
29
 5
5
88
29
134
 1.1
1.1
18
14
24
 6
6
98
123
826
 10
10
171
26
317
 13
13
229
94
355
 14
14
239
109
299
 10
10
173
29
240
 3
3
51
34
106
 5
5
87
20
110
 4
4
70
26
90
 5
5
83
28
135
 50
50
849
65
1835
 3
3
50
43
86
 7
7
125
26
184
 5
5
85
25
108
 4
4
73
28
58
 14
14
243
24
247
 100
100
1701
36
3135
 35
35
594
70
1259
 46
46
776
52
695
 6
6
103
35
116
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 94.4
94.4
96
98 88.5
88.5
95.2
- -
-
-
100 -
-
-
- 99.6
99.6
99.8
100 -
-
-
- 83.6
83.6
90.2
100 99.8
99.8
100
100 -
-
-
- 73.8
73.8
74.6
- 87.1
87.1
94
98 86.6
86.6
94.3
96.5 84.1
84.1
90.8
94 -
-
-
- 70.4
70.4
80.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
37.5 30.5
30.5
47.6
41 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | GRIN2B - Q13224 |
| 2 | LRRC7 - Q96NW7 |
| 3 | ATP2A2 - P16615 |
| 4 | ETS1 - P14921 |
| 5 | GRIN2A - Q12879 |
| 6 | PPM1F - P49593 |
| 7 | ITGB1BP1 - O14713 |
| 8 | DAPK2 - Q9UIK4 |
| 9 | CHAT - P28329 |
| 10 | HSF1 - Q00613 |
| 11 | CAMK2N2 - Q96S95 |
| 12 | EGFR - P00533 |
| 13 | SYNGAP1 - Q96PV0 |
| 14 | PDC - P20941 |
| 15 | CDK5R2 - Q13319 |
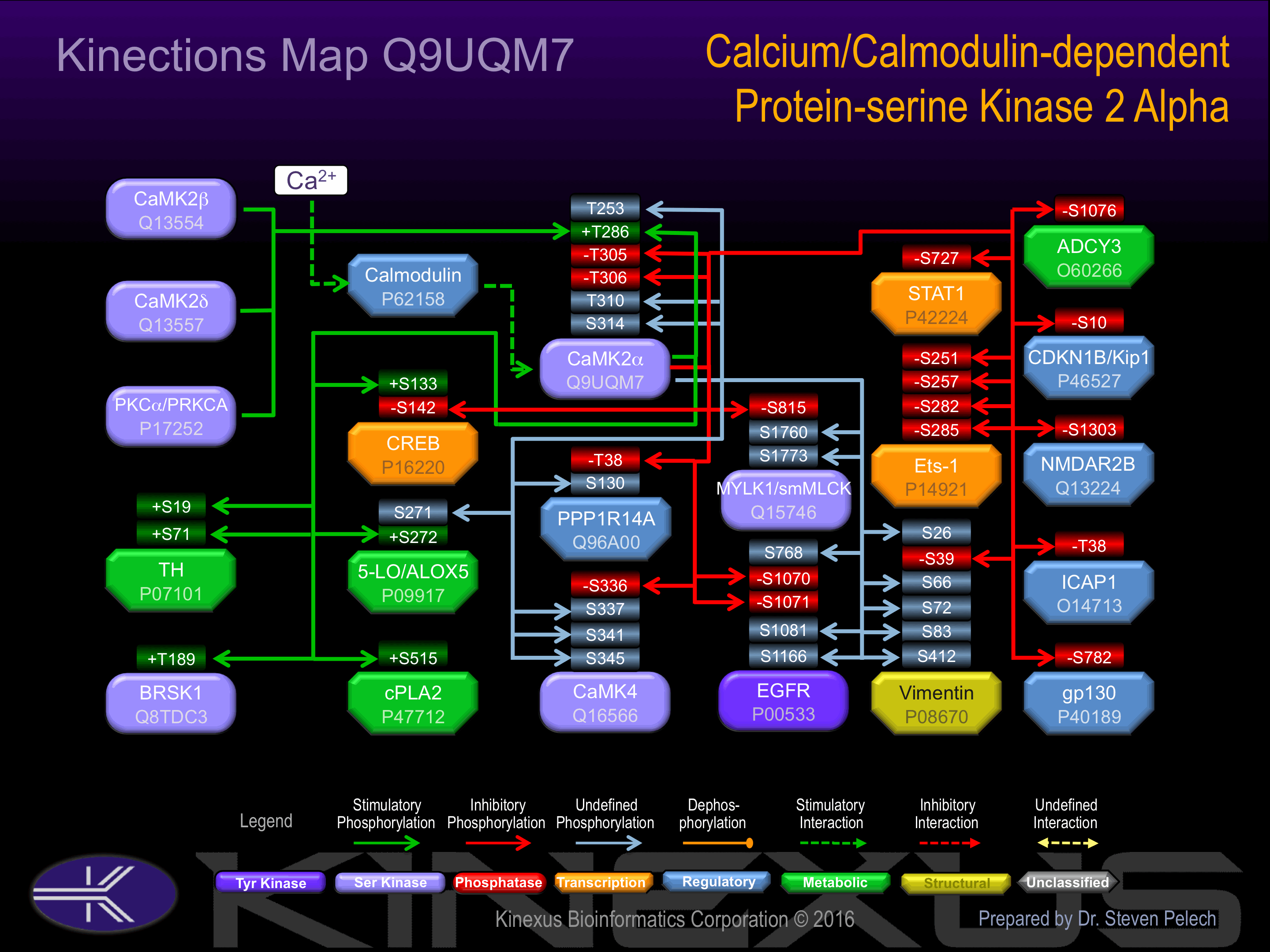
Regulation
Activation:
Autophosphorylation of Thr-286 allows the kinase to switch from a calmodulin-dependent to a calmodulin-independent state
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CaMK2a | Q9UQM7 | T253 | DLINKMLTINPSKRI | ? |
| CaMK2a | Q9UQM7 | S279 | SHRSTVASCMHRQET | |
| CaMK2b | Q13554 | T286 | SCMHRQETVDCLKKF | + |
| CaMK2d | Q13557 | T286 | SCMHRQETVDCLKKF | + |
| PKCa | P17252 | T286 | SCMHRQETVDCLKKF | + |
| CaMK2a | Q9UQM7 | T286 | SCMHRQETVDCLKKF | + |
| CaMK2a | Q9UQM7 | T305 | KLKGAILTTMLATRN | - |
| CaMK2a | Q9UQM7 | T306 | LKGAILTTMLATRNF | - |
| CaMK2a | Q9UQM7 | T310 | ILTTMLATRNFSGGK | |
| CaMK2b | Q13554 | S314 | MLATRNFSGGKSGGN | |
| CaMK2a | Q9UQM7 | S314 | MLATRNFSGGKSGGN |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| 14-3-3 beta (YWHAB) | P31946 | S59 | NVVGARRSSWRVISS | |
| 14-3-3 beta (YWHAB) | P31946 | S64 | RSSWRVISSIEQKTE | |
| 14-3-3 beta (YWHAB) | P31946 | S65 | SSWRVISSIEQKTER | |
| 5-LO | P09917 | S271 | CSLERQLSLEQEVQQ | |
| 5-LO (ALOX5) | P09917 | S272 | CSLERQLSLEQEVQQ | + |
| ACC1 | Q13085 | S25 | RFIIGSVSEDNSEDE | |
| ADCY3 | O60266 | S1076 | NVASRMESTGVMGNI | - |
| APP | P05067 | T729 | MLKKKQYTSIHHGVV | |
| ASIC1 | P78348 | S524 | CQKEAKRSSADKGVA | |
| ASIC1 | P78348 | S525 | QKEAKRSSADKGVAL | |
| BRSK1 | Q8TDC3 | T189 | VGDSLLETSCGSPHY | + |
| CACNA1B | Q00975 | S2129 | SEKQRFYSCDRFGGR | |
| CACNA1B | Q00975 | S783 | RLQNLRASCEALYSE | |
| CACNA1B | Q00975 | S891 | ERPRPHRSHSKEAAG | |
| CACNA1C | Q13936 | S1535 | DYLTRDWSILGPHHL | |
| CACNA1C | Q13936 | S1593 | VACKRLVSMNMPLNS | |
| CACNA1C | Q13936 | S409 | GVLSGEFSKEREKAK | |
| CACNA1C | Q13936 | T1622 | RTALRIKTEGNLEQA | |
| CACNB2 | Q08289 | T554 | RGLSRQETFDSETQE | |
| Caldesmon | Q05682 | S26 | RLEAERLSYQRNDDD | |
| Caldesmon | Q05682 | S58 | RQKQEEESLGQVTDQ | |
| Caldesmon | Q05682 | S643 | CFTPKGSSLKIEERA | |
| Caldesmon | Q05682 | S677 | AIVSKIDSRLEQYTS | |
| Caldesmon | Q05682 | S73 | VEVNAQNSVPDEEAK | |
| Caldesmon | Q05682 | S783 | RNLWEKQSVDKVTSP | |
| Calponin 1 | P51911 | S175 | MGTNKFASQQGMTAY | |
| CaMK2a | Q9UQM7 | S279 | SHRSTVASCMHRQET | |
| CaMK2a | Q9UQM7 | S314 | MLATRNFSGGKSGGN | |
| CaMK2a | Q9UQM7 | T253 | DLINKMLTINPSKRI | ? |
| CaMK2a | Q9UQM7 | T286 | SCMHRQETVDCLKKF | + |
| CaMK2a | Q9UQM7 | T305 | KLKGAILTTMLATRN | - |
| CaMK2a | Q9UQM7 | T306 | LKGAILTTMLATRNF | - |
| CaMK2a | Q9UQM7 | T310 | ILTTMLATRNFSGGK | |
| CaMK4 | Q16566 | S336 | AVKAVVASSRLGSAS | - |
| CaMK4 | Q16566 | S337 | VKAVVASSRLGSASS | |
| CaMK4 | Q16566 | S341 | VASSRLGSASSSHGS | |
| CaMK4 | Q16566 | S345 | RLGSASSSHGSIQES | |
| CARD11 | Q9BXL7 | S116 | KEPTRRFSTIVVEEG | |
| CD44 | P16070 | S706 | LNGEASKSQEMVHLV | |
| ChAT | P28329 | T574 | VDNIRSATPEALAFV | |
| CLC-3 | P51790 | S51 | ERHRRINSKKKESAW | |
| CPEB | Q9BZB8 | T172 | VRGSRLDTRPILDSR | |
| cPLA2 | P47712 | S515 | SDFATQDSFDDDELD | + |
| CREB1 | P16220 | S133 | EILSRRPSYRKILND | + |
| CREB1 | P16220 | S142 | RKILNDLSSDAPGVP | - |
| CREM | Q03060-2 | S71 | EILSRRPSYRKILNE | |
| DAT (Dopamine transporter) | Q01959 | S12 | KCSVGLMSSVVAPAK | |
| DAT (Dopamine transporter) | Q01959 | S13 | CSVGLMSSVVAPAKE | |
| DAT (Dopamine transporter) | Q01959 | S2 | ______MSKSKCSVG | |
| DAT (Dopamine transporter) | Q01959 | S4 | ____MSKSKCSVGLM | |
| DAT (Dopamine transporter) | Q01959 | S7 | _MSKSKCSVGLMSSV | |
| DLG1 (SAP97) | Q12959 | S232 | ITLERGNSGLGFSIA | |
| DLG4 (PSD-95) | P78352 | S73 | ITLERGNSGLGFSIA | |
| DRD3 | P35462 | S229 | RILTRQNSQCNSVRP | |
| EGFR | P00533 | S1070 | DSFLQRYSSDPTGAL | - |
| EGFR | P00533 | S1071 | SFLQRYSSDPTGALT | - |
| EGFR | P00533 | S1081 | TGALTEDSIDDTFLP | |
| EGFR | P00533 | S1166 | QKGSHQISLDNPDYQ | |
| EGFR | P00533 | S768 | DEAYVMASVDNPHVC | |
| ErbB2 (HER2) | P04626 | T1172 | ATLERPKTLSPGKNG | ? |
| Ets-1 | P14921 | S251 | GKLGGQDSFESIESY | - |
| Ets-1 | P14921 | S257 | DSFESIESYDSCDRL | - |
| Ets-1 | P14921 | S282 | NSLQRVPSYDSFDSE | - |
| Ets-1 | P14921 | S285 | QRVPSYDSFDSEDYP | - |
| Ets-2 | P15036 | S246 | FPKSRLSSVSVTYCS | |
| Ets-2 | P15036 | S310 | LDVQRVPSFESFEDD | |
| Ets-2 | P15036 | S313 | QRVPSFESFEDDCSQ | |
| FLNA | P21333 | S2523 | VTGPRLVSNHSLHET | |
| GABRA-G2 | P18507 | S348 | VCFIFVFSALVEYGT | |
| GABRB1 | P18505 | S409 | IQYRKPLSSREAYGR | |
| GABRB1 | P18505 | S434 | GRIRRRASQLKVKIP | |
| GABRG2 | P18507 | S386 | TIDIRPRSATIQMNN | |
| GABRG2 | P18507 | T388 | DIRPRSATIQMNNAT | |
| GLO1 | Q04760 | T107 | ELTHNWGTEDDETQS | |
| GluR1 | P42261 | S645 | LTVERMVSPIESAED | |
| GluR1 | P42261 | S849 | FCLIPQQSINEAIRT | |
| GluR4 | P48058 | S862 | IRNKARLSITGSVGE | |
| gp130 | P40189 | S782 | QVFSRSESTQPLLDS | - |
| HSF1 | Q00613 | S230 | PKYSRQFSLEHVHGS | |
| HSL | Q05469 | S554 | EPMRRSVSEAALAQP | |
| ICAP1 | O14713 | T38 | GGLSRSSTVASLDTD | - |
| ITGB1 | P05556 | T788 | PIYKSAVTTVVNPKY | |
| ITGB1 | P05556 | T789 | IYKSAVTTVVNPKYE | |
| ITPKA | P23677 | T311 | EHAQRAVTKPRYMQW | |
| KIF17 | Q9P2E2 | S1020 | TKAKRKKSKSNFGSE | |
| KRT8 | P05787 | S432 | SAYGGLTSPGLSYSL | |
| Kv4.2 | Q9NZV8 | S438 | ARIRAAKSGSANAYM | |
| Kv4.2 | Q9NZV8 | S459 | LLSNQLQSSEDEQAF | |
| Kv4.3 | Q9UK17 | S569 | LPATRLRSMQELSTI | |
| LRP4 | O75096 | S1932 | ATPERRGSLPDTGWK | |
| LRP4 | O75096 | S1945 | WKHERKLSSESQV__ | |
| MeCP2 | P51608 | S423 | EKMPRGGSLESDGCP | |
| MOR1 | P35372 | S263 | LMILRLKSVRMLSGS | |
| MOR1 | P35372 | S268 | LKSVRMLSGSKEKDR | |
| myelin P0 | P25189 | S210 | HKPGKDASKRGRQTP | |
| myelin P0 | P25189 | S233 | SRSTKAVSEKKAKGL | |
| MYLK1 (smMLCK) | Q15746 | S1760 | RAIGRLSSMAMISGL | |
| MYLK1 (smMLCK) | Q15746 | S1773 | GLSGRKSSTGSPTSP | |
| MYLK1 (smMLCK) | Q15746 | S815 | SLMLQNSSARALPRG | - |
| Myogenin | P15173 | T87 | VDRRRAATLREKRRL | |
| NeuroD | Q13562 | S335 | IPIDNIMSFDSHSHH | |
| NFL (Neurofilament L) | P07196 | S54 | SSLSVRRSYSSSSGS | |
| NFL (Neurofilament L) | P07196 | S69 | SLMPSLESLDLSQVA | |
| NMDAR2B (GRIN2B) | Q13224 | S1303 | NKLRRQHSYDTFVDL | - |
| nNOS | P29475 | S847 | SYKVRFNSVSSYSDS | |
| NUMB | P49757 | S276 | EQLARQGSFRGFPAL | |
| p27Kip1 | P46527 | S10 | NVRVSNGSPSLERMD | - |
| PEA-15 | Q15121 | S116 | KDIIRQPSEEEIIKL | |
| Phosducin | P20941 | S54 | KEILRQMSSPQSRNG | |
| Phosducin | P20941 | S73 | ERVSRKMSIQEYELI | |
| Phospholamban | P26678 | S16 | RSAIRRASTIEMPQQ | |
| Phospholamban | P26678 | T17 | SAIRRASTIEMPQQA | |
| PLCB3 | Q01970 | S537 | PSLEPQKSLGDEGLN | |
| PPP1R14A (CPI 17) | Q96A00 | S130 | GLRQPSPSHDGSLSP | |
| PPP1R14A (CPI 17) | Q96A00 | T38 | QKRHARVTVKYDRRE | - |
| PPP1R3A | Q16821 | S46 | PQPSRRGSDSSEDIY | |
| Rabphilin 3A | Q9Y2J0 | S272 | AGLRRANSVQASRPA | |
| Rabphilin 3A | Q9Y2J0 | S34 | EQLQAGWSVHPGGQP | |
| RIMS1 | Q86UR5 | S218 | RLQERSRSQTPLSTA | |
| RIMS1 | Q86UR5 | S265 | QASSRSRSEPPRERK | |
| RRAD | P55042 | S273 | AGTRRRESLGKKAKR | |
| RyR2 | Q92736 | S2806 | YNRTRRISQTSQVSV | |
| Separase | Q14674 | S1501 | TDNWRKMSFEILRGS | |
| SERCA2 | P16615 | S38 | KLKERWGSNELPAEE | |
| Smad2 | Q15796 | S110 | SFSEQTRSLDGRLQV | |
| Smad2 | Q15796 | S240 | SDQQLNQSMDTGSPA | |
| Smad2 | Q15796 | S260 | TLSPVNHSLDLQPVT | |
| SNCA | P37840 | S129 | NEAYEMPSEEGYQDY | |
| Spinophilin | Q96SB3 | S100 | LSLPRASSLNENVDH | |
| SPR | P35270 | S213 | QQLARETSVDPDMRK | |
| SRF | P11831 | S103 | RGLKRSLSEMEIGMV | |
| STAT1 | P42224 | S727 | TDNLLPMSPEEFDEV | - |
| STMN1 | P16949 | S16 | KELEKRASGQAFELI | |
| SYN1 | P17600 | S568 | PQATRQTSVSGPAPP | |
| SYN1 | P17600 | S605 | AGPTRQASQAGPVPR | |
| synGAP | Q96PV0 | S1138 | PSITKQHSQTPSTLN | |
| synGAP | Q96PV0 | S780 | MARGLNSSMDMARLP | |
| SYT1 | P21579 | T113 | DVKDLGKTMKDQALK | |
| Tau iso5 (Tau-C) | P10636-5 | S131 | QARMVSKSKDGTGSD | |
| Tau iso5 (Tau-C) | P10636-5 | S214 | GSRSRTPSLPTPPTR | |
| Tau iso5 (Tau-C) | P10636-5 | S262 | NVKSKIGSTENLKHQ | |
| Tau iso5 (Tau-C) | P10636-5 | T135 | VSKSKDGTGSDDKKA | |
| Tau iso5 (Tau-C) | P10636-5 | T212 | TPGSRSRTPSLPTPP | |
| Tau iso9 (Tau-F) | P10636-9 | S356 | RVQSKIGSLDNITHV | |
| Tau iso9 (Tau-F) | P10636-9 | S409 | GTSPRHLSNVSSTGS | |
| Tau iso9 (Tau-F) | P10636-9 | S416 | SNVSSTGSIDMVDSP | |
| TH | P07101 | S19 | KGFRRAVSEQDAKQA | + |
| TH | P07101 | S71 | RFIGRRQSLIEDARK | + |
| TH iso3 | P07101-3 | S35 | AIMVRGQSPRFIGRR | |
| TPH2 | Q8IWU9 | S19 | YWARRGFSLDSAVPE | |
| TRAD | O60229 | T95 | DVCKRGFTVIIDMRG | |
| VAMP1 | P23763 | S63 | LERDQKLSELDDRAD | |
| Vimentin | P08670 | S26 | GTASRPSSSRSYVTT | |
| Vimentin | P08670 | S39 | TTSTRTYSLGSALRP | - |
| Vimentin | P08670 | S412 | EGEESRISLPLPNFS | |
| Vimentin | P08670 | S66 | GVYATRSSAVRLRSS | |
| Vimentin | P08670 | S72 | SSAVRLRSSVPGVRL | |
| Vimentin | P08670 | S83 | GVRLLQDSVDFSLAD | |
| VR1 | Q8NER1 | S502 | YFLQRRPSMKTLFVD | |
| VR1 | Q8NER1 | T705 | WKLQRAITILDTEKS |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
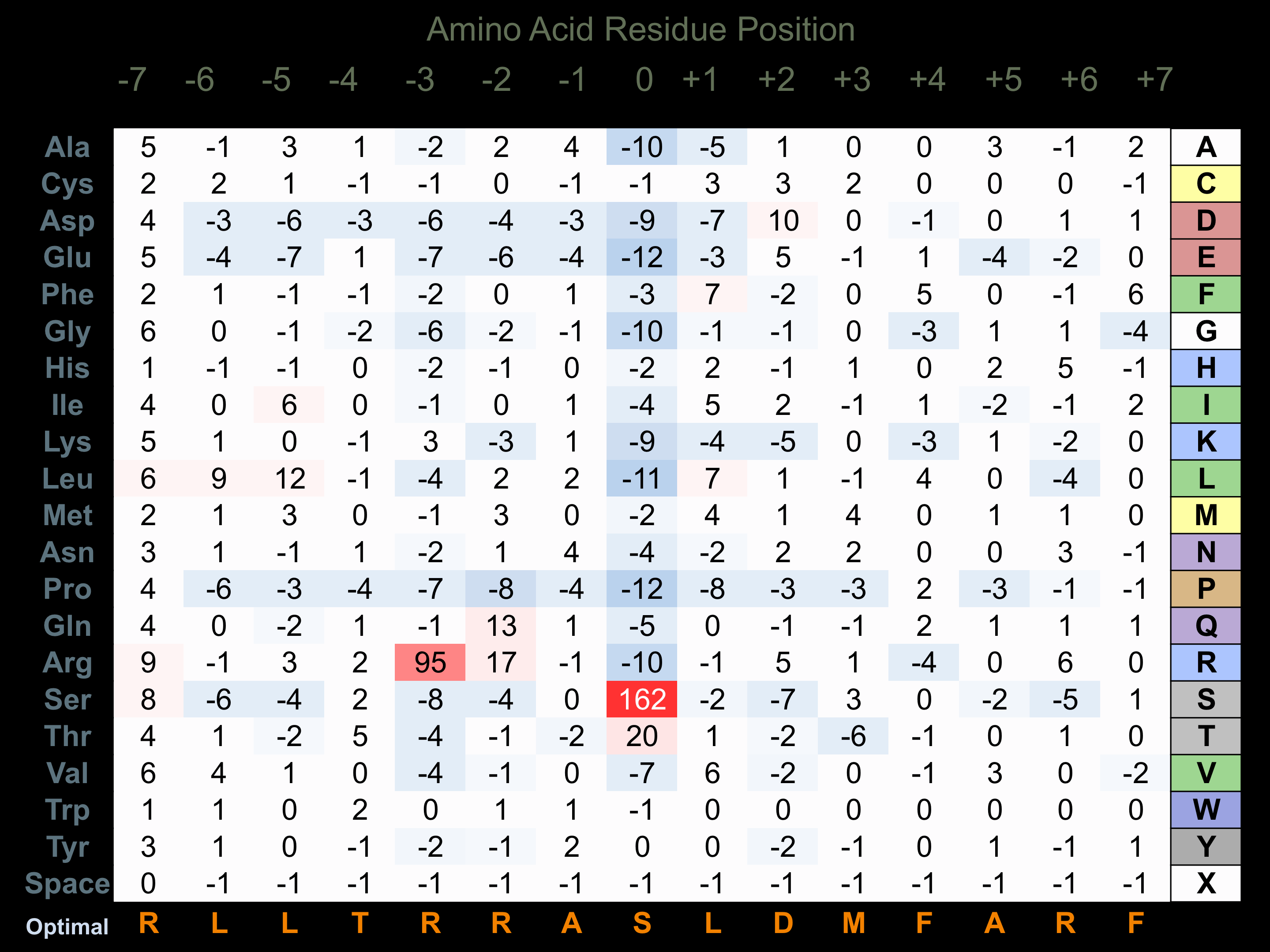
Matrix Type:
Experimentally derived from alignment of 302 known protein substrate phosphosites and 31 peptides phosphorylated by recombinant CaMK2-alpha in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Heart disorders
Specific Diseases (Non-cancerous):
Arrhythmogenesis
Comments:
O-GlcNAc binding to CaMK2a at S279 will result in activation, which can lead to an arrhythmia of the heart. CaMK2I inhibition may substantially reduce maladaptive remodeling from excessive beta-adrenergic receptor stimulation and myocardial infarction, and induce balanced changes in excitation-contraction coupling that preserve baseline and beta-adrenergic rceeptor-stimulated physiological increases in cardiac function.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in Bladder carcinomas (%CFC= -55, p<0.012). The COSMIC website notes an up-regulated expression score for CaMK2a in diverse human cancers of 240, which is 48% lower than the average score of 462 for the human protein kinases. The down-regulated expression score of 1 for this protein kinase in human cancers was 98% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24727 diverse cancer specimens. This rate is only 13 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.57 % in 589 stomach cancers tested; 0.39 % in 864 skin cancers tested; 0.31 % in 603 endometrium cancers tested; 0.3 % in 1270 large intestine cancers tested; 0.23 % in 273 cervix cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,010 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.