Nomenclature
Short Name:
CaMK2b
Full Name:
Calcium-calmodulin-dependent protein kinase type II beta chain
Alias:
- Calcium/calmodulin-dependent protein kinase II beta
- Calcium/calmodulin-dependent protein kinase type II beta chain
- CAMKB
- CaMK-II beta
- CaMK-II beta subunit
- Proline rich calmodulin-dependent protein kinase; CaM-kinase II beta chain; EC 2.7.1.123; EC 2.7.11.17; KCC2B; Kinase CaMK2-beta;
- CaM kinase II beta subunit
- CAM2
- CAMK2
- CaMK2-beta
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
CAMK2
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
72,727
# Amino Acids:
664
# mRNA Isoforms:
8
mRNA Isoforms:
72,678 Da (666 AA; Q13554); 60,387 Da (542 AA; Q13554-2); 58,009 Da (518 AA; Q13554-5); 57,938 Da (517 AA; Q13554-8); 56,379 Da (503 AA; Q13554-3); 55,223 Da (492 AA; Q13554-6); 54,074 Da (479 AA; Q13554-4); 50,955 Da (449 AA; Q13554-7)
4D Structure:
CAMK2 is composed of four different chains: alpha, beta, gamma, and delta. The different isoforms assemble into homo- or heteromultimeric holoenzymes composed of 8 to 12 subunits. Interacts with SYNGAP1 and CAMK2N2.Interacts with MPDZ.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
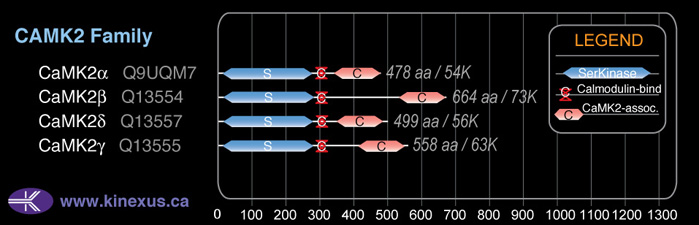
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 14 | 272 | Pkinase |
| 297 | 312 | CaM_binding |
| 532 | 659 | CaMK2-Association |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S79, S146, S235, S276, S280, S315, S394, S395, S397, S423, S436, S441, S456, S461, S471, S479, S499, S504, S522.
Threonine phosphorylated:
T254, T287+, T306, T307, T311, T328, T400, T401, T463, T506.
Tyrosine phosphorylated:
Y231.
Ubiquitinated:
K227, K246, K268.
Acetylated:
K227, K342.
Methylated:
K301.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 64
64
624
71
956
 8
8
77
38
85
 13
13
130
24
289
 100
100
981
215
646
 52
52
511
61
426
 2
2
24
185
42
 16
16
161
77
456
 78
78
765
86
683
 25
25
242
44
192
 12
12
113
176
254
 8
8
79
71
94
 60
60
588
385
559
 5
5
50
79
95
 4
4
44
29
47
 16
16
159
56
446
 8
8
76
37
80
 7
7
73
250
74
 9
9
85
47
179
 35
35
339
224
300
 24
24
237
274
274
 23
23
226
51
215
 6
6
59
61
91
 9
9
90
29
151
 9
9
85
47
119
 8
8
80
51
208
 71
71
697
128
1153
 6
6
57
88
93
 9
9
85
47
151
 7
7
69
47
118
 29
29
282
70
398
 21
21
202
42
294
 88
88
860
81
1962
 28
28
271
154
770
 72
72
710
130
613
 18
18
177
83
228
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 66.8
66.8
77.3
0 62.6
62.6
63.4
- -
-
-
94.5 -
-
-
- 74.5
74.5
82
- -
-
-
- 81
81
81.3
98 80.9
80.9
81.2
98 -
-
-
- -
-
-
- 64
64
68.8
92 63.4
63.4
68.1
90 69.9
69.9
76.2
87 -
-
-
- 60.1
60.1
68.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
40 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CAMK2N2 - Q96S95 |
| 2 | GAPDH - P04406 |
| 3 | PLCB3 - Q01970 |
| 4 | COL4A3 - Q01955 |
| 5 | CAMK2D - Q13557 |
| 6 | GRIN2A - Q12879 |
| 7 | ETS1 - P14921 |
| 8 | GRIN2B - Q13224 |
| 9 | ACTN4 - O43707 |
| 10 | ITPKA - P23677 |
| 11 | CAMK2A - Q9UQM7 |
| 12 | GRIN1 - Q05586 |
Regulation
Activation:
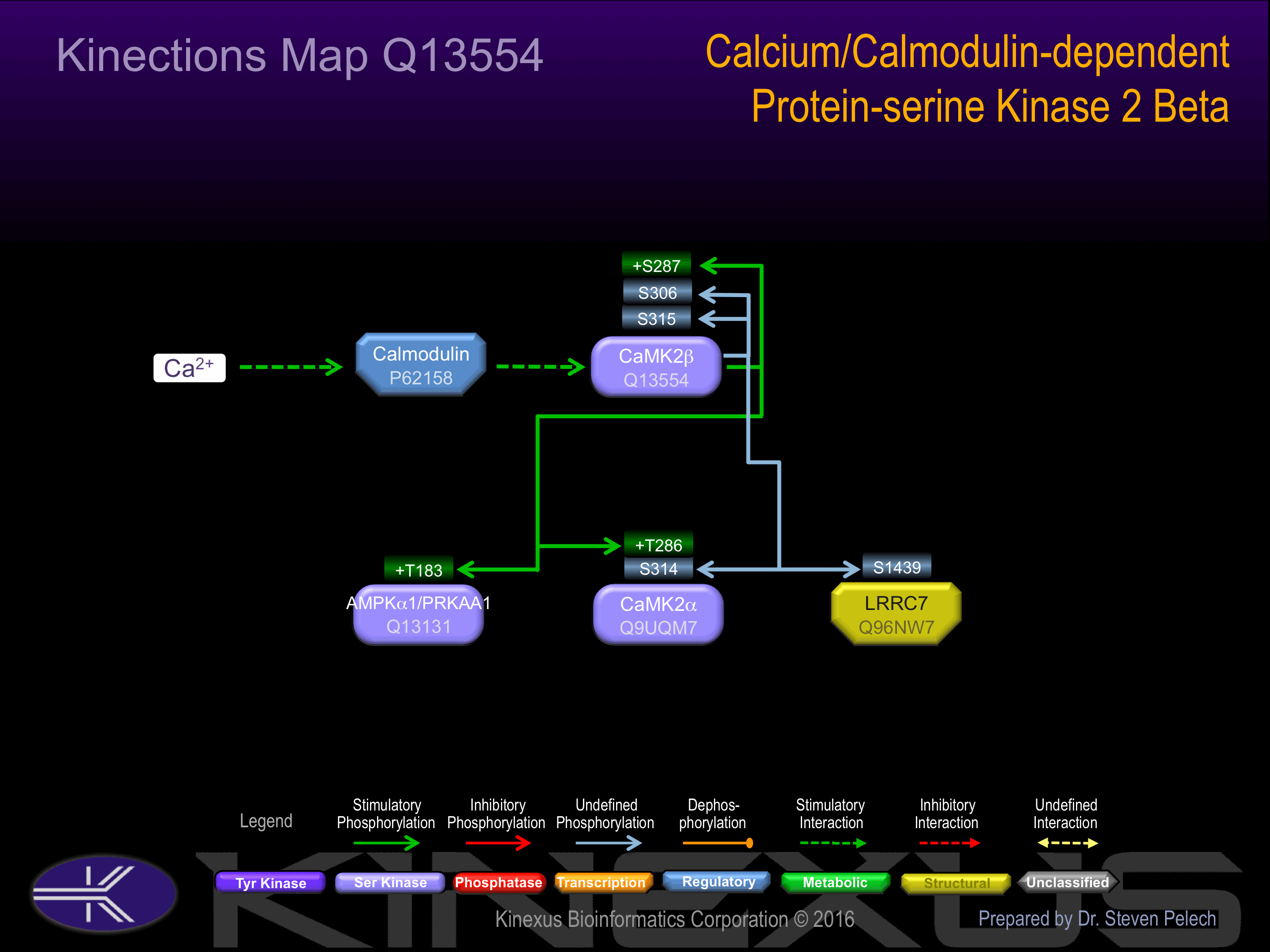
Activated in response to a rise in intracellular Ca(2+) concentration. Autophosphorylation at Thr-287 increases phosphotransferase activity in a Ca(2+)-independent manner.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
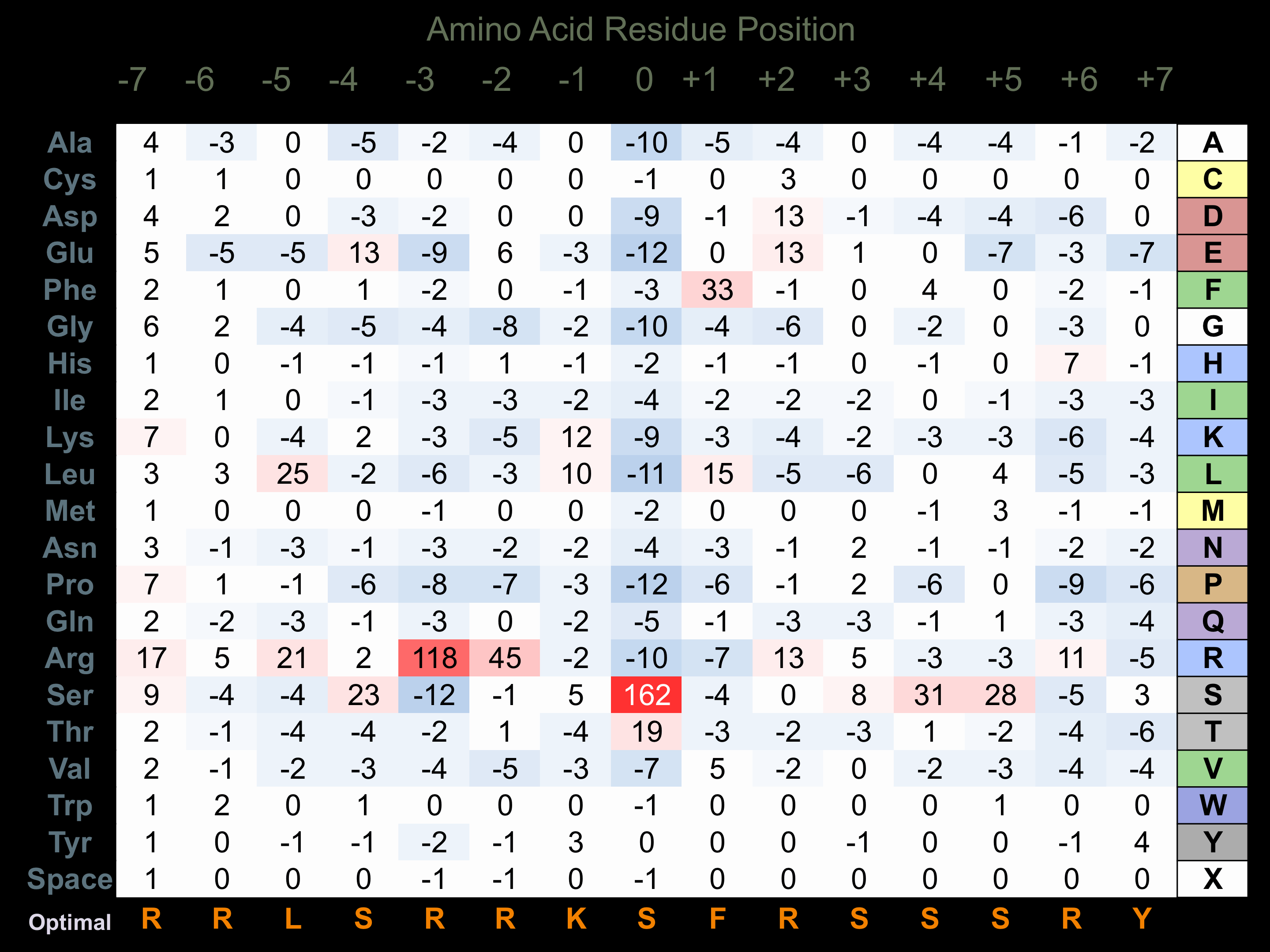
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Phencyclidine abuse
Comments:
One of the diseases associated with this protein kinase include phencyclidine abuse.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Pituitary adenomas (ACTH-secreting) (%CFC= +228, p<0.057). The COSMIC website notes an up-regulated expression score for CAMK2b in diverse human cancers of 356, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24846 diverse cancer specimens. This rate is only 17 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.45 % in 805 skin cancers tested; 0.32 % in 1093 large intestine cancers tested; 0.28 % in 589 stomach cancers tested; 0.26 % in 934 upper aerodigestive tract cancers tested; 0.15 % in 1619 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: G468A (9).
Comments:
16 insertions (14 at C477fs*8 that result in a truncated protein with loss of calcium-calmodulin binding domain), 2 deletionsand 2 complex mutations are noted on the COSMIC website.