Nomenclature
Short Name:
DDR1
Full Name:
Epithelial discoidin domain receptor 1
Alias:
- CAK
- CD167
- Discoidin receptor tyrosine kinase
- EC 2.7.1.112
- EC 2.7.10.1
- NEP; NTRK4; PTK3; PTK3A; RTK6; TRKE; Tyrosine- protein kinase CAK
- CD167a
- CD167a antigen
- Cell adhesion kinase
- Discoidin domain receptor tyrosine kinase 1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
DDR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
101,128
# Amino Acids:
913
# mRNA Isoforms:
5
mRNA Isoforms:
101,796 Da (919 AA; Q08345-5); 101,128 Da (913 AA; Q08345); 99,064 Da (894 AA; Q08345-6); 97,174 Da (876 AA; Q08345-2); 27,130 Da (243 AA; Q08345-4)
4D Structure:
Interacts (via PPxY motif) with WWC1 (via WW domains) in a collagen-regulated manner. Forms a tripartite complex with WWC1 and PRKCZ, but predominantly in the absence of collagen.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 20 | signal_peptide |
| 31 | 185 | FA58C |
| 417 | 439 | TMD |
| 610 | 905 | TyrKc |
| 610 | 907 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K177.
Methylated:
K613, K615.
N-GlcNAcylated:
N211, N260, N370, N371, N394.
Serine phosphorylated:
S45, S496, S631, S788.
Threonine phosphorylated:
T469, T519.
Tyrosine phosphorylated:
Y484, Y513, Y520, Y703, Y740, Y792+, Y796+, Y797+, Y869, Y881.
Ubiquitinated:
K112, K153, K449.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 65
65
1003
55
921
 24
24
372
27
248
 13
13
206
51
308
 58
58
901
206
681
 100
100
1541
47
736
 27
27
418
148
168
 42
42
643
59
497
 58
58
899
104
1679
 71
71
1100
31
618
 28
28
426
190
295
 10
10
157
85
250
 61
61
933
349
756
 10
10
152
95
199
 11
11
173
21
123
 24
24
368
74
487
 24
24
369
30
166
 28
28
435
264
265
 20
20
313
66
337
 17
17
258
208
194
 69
69
1068
215
759
 52
52
809
70
715
 12
12
178
77
274
 17
17
269
63
257
 10
10
160
66
231
 20
20
302
70
375
 61
61
946
138
760
 14
14
209
98
168
 30
30
466
66
404
 15
15
228
66
327
 28
28
425
56
237
 25
25
391
36
481
 46
46
708
66
637
 0.2
0.2
3
68
1
 49
49
753
109
653
 18
18
283
70
285
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.6
100 95.3
95.3
95.5
99 -
-
-
96 -
-
-
98 95.1
95.1
96.6
96 -
-
-
- 93.7
93.7
95.8
94 93.8
93.8
96
95 -
-
-
- 53.5
53.5
65.9
- 24.5
24.5
41.5
- 60.6
60.6
72.4
64 52.3
52.3
65.7
64 -
-
-
- 23.6
23.6
40.3
- 34.4
34.4
50.6
- -
-
-
- 39.1
39.1
54.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
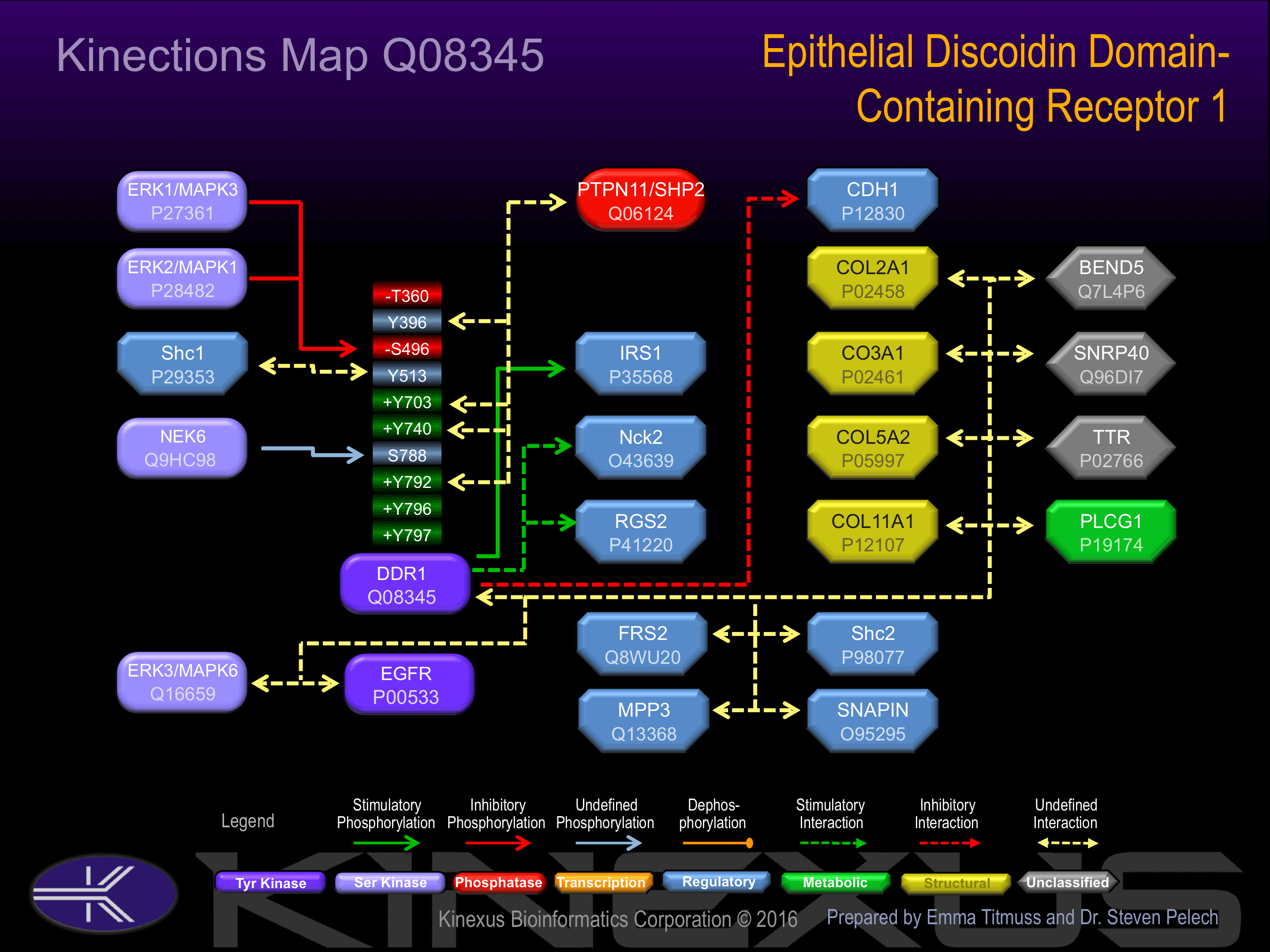
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | IRS1 - P35568 |
| 2 | SHC1 - P29353 |
| 3 | FRS2 - Q8WU20 |
| 4 | PTPN11 - Q06124 |
| 5 | NCK2 - O43639 |
| 6 | COL3A1 - P02461 |
Regulation
Activation:
Autophosphorylated in response to fibrillar collagen binding. Phosphorylation at Tyr-396, Tyr-703 and Tyr-796 induces interaction with SHP2 and Y513 to Shc1.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
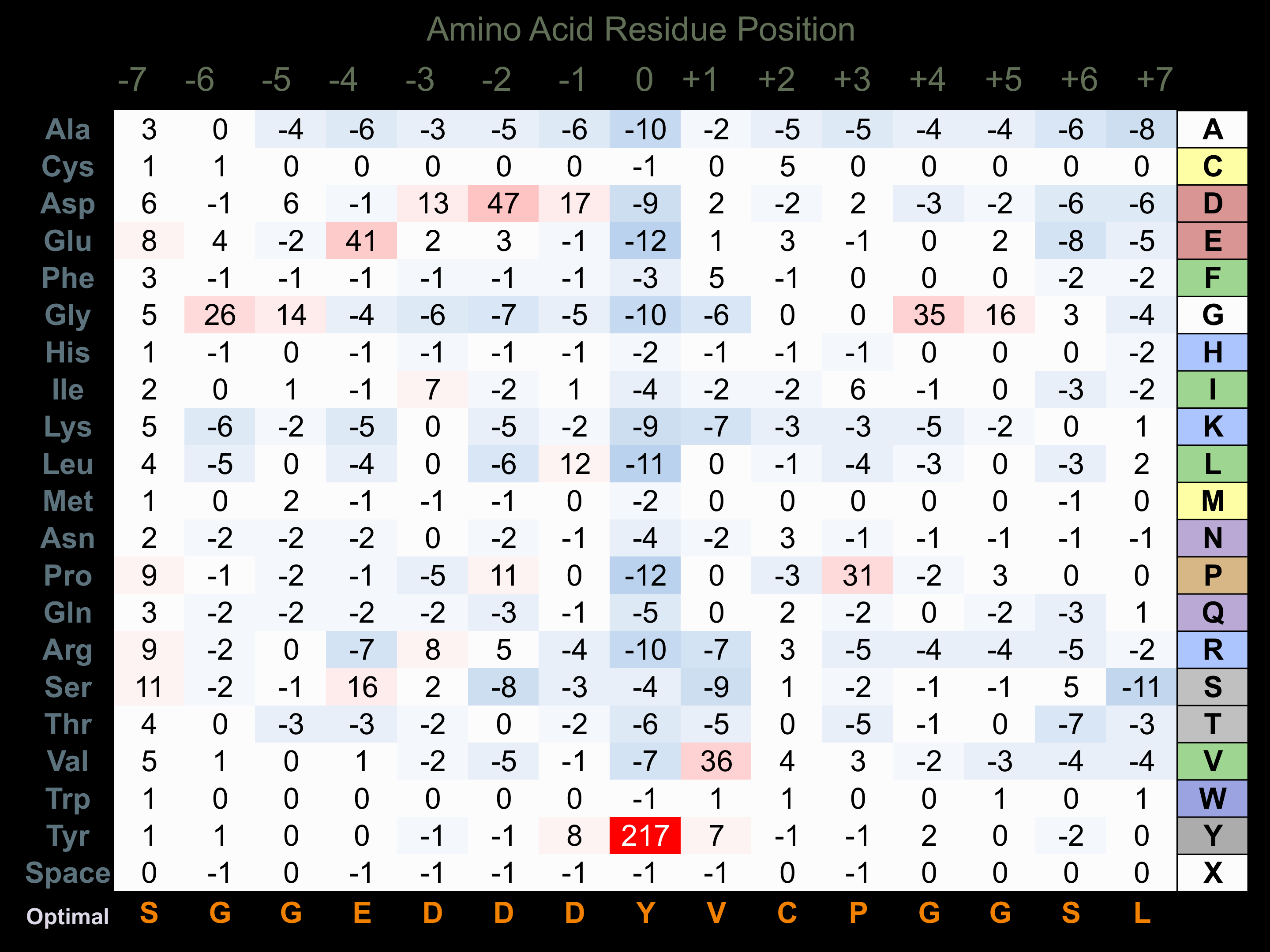
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Osteosarcoma
Comments:
DDR1 may be an oncoprotein (OP) or a tumour suppressor protein (TSP). DDR1 has been shown to be significantly overexpressed in some human tumours. Multiple sites of mutation lead to either inhibition or constitutively activation of this receptor. The gene expression was found to be upregulated by p53 in human osteosarcoma cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +122, p<0.0001); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +107, p<0.0001); Ovary adenocarcinomas (%CFC= +99, p<0.0001); Pituitary adenomas (ACTH-secreting) (%CFC= -50, p<0.013); Pituitary adenomas (aldosterone-secreting) (%CFC= +49, p<0.0001); Skin fibrosarcomas (%CFC= -78, p<0.0001); Skin melanomas (%CFC= -61, p<0.0001); and T-cell prolymphocytic leukemia (%CFC= +316, p<0.0001). The COSMIC website notes an up-regulated expression score for DDR1 in diverse human cancers of 458, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 75 for this protein kinase in human cancers was 1.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25589 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.33 % in 805 skin cancers tested; 0.32 % in 1093 large intestine cancers tested; 0.24 % in 589 stomach cancers tested; 0.1 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,842 cancer specimens
Comments:
Only 5 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.