Nomenclature
Short Name:
DMPK2
Full Name:
Serine-threonine-protein kinase MRCK gamma
Alias:
- EC 2.7.11.1
- MRCKG
- HSMDPKIN
- Myotonic dystrophy protein kinase like protein
- CDC42BPG
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
DMPK
SubFamily:
GEK
Structure
Mol. Mass (Da):
172459
# Amino Acids:
1551
# mRNA Isoforms:
1
mRNA Isoforms:
172,459 Da (1551 AA; Q6DT37)
4D Structure:
Homodimer and homotetramer via the coiled coil regions. Interacts tightly with GTP-bound but not GDP-bound CDC42
1D Structure:
Subfamily Alignment
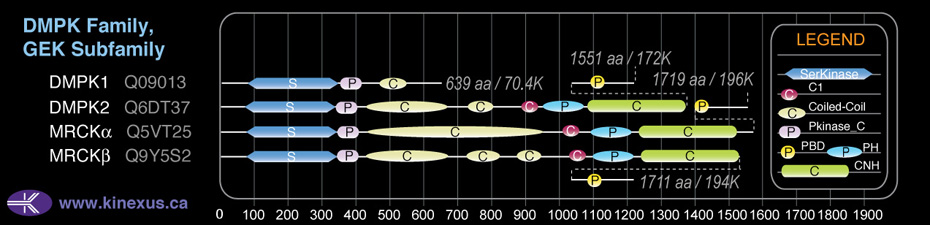
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 71 | 337 | Pkinase |
| 338 | 408 | Pkinase_C |
| 406 | 678 | Coiled-coil |
| 730 | 802 | Coiled-coil |
| 878 | 927 | C1 |
| 947 | 1066 | PH |
| 1092 | 1366 | CNH |
| 1437 | 1450 | PBD |
| 1437 | 1450 | CRIB |
| 744 | 801 | DMPK coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S239, S480, S740, S747, S817, S851, S1121, S1482, S1484, S1492, S1514, S1515, S1540.
Threonine phosphorylated:
T222, T234.
Ubiquitinated:
K75, K125, K663, K1172, K1385.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1332
15
1468
 4
4
49
7
58
 -
-
-
-
-
 3
3
34
51
59
 38
38
505
17
331
 0.8
0.8
10
18
8
 0.9
0.9
12
23
8
 6
6
79
5
110
 1.1
1.1
15
3
2
 7
7
89
5
115
 5
5
68
5
80
 39
39
524
28
539
 4
4
54
2
17
 7
7
87
5
125
 8
8
111
5
174
 3
3
41
12
47
 2
2
22
13
42
 14
14
188
5
329
 14
14
184
5
237
 29
29
382
51
156
 3
3
42
5
39
 0.5
0.5
6
3
3
 -
-
-
-
-
 7
7
95
7
106
 3
3
34
5
36
 42
42
564
37
512
 4
4
55
5
64
 8
8
102
5
113
 11
11
152
5
261
 -
-
-
-
-
 44
44
580
12
35
 7
7
94
12
29
 25
25
328
57
531
 57
57
760
52
657
 3
3
41
35
27
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 90.3
90.3
90.8
99 94.5
94.5
95.7
97 -
-
-
88 -
-
-
- 87.3
87.3
91.7
88 -
-
-
- 85
85
89.7
85 45
45
62.2
85 -
-
-
- 45.9
45.9
62.6
- 46.4
46.4
63.8
- -
-
-
51 37.1
37.1
52.3
- -
-
-
- 37.5
37.5
56.1
- 37.3
37.3
54.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Agonist binding to the phorbol ester binding site disrupts this, releasing the kinase domain to allow N-terminus-mediated dimerization and kinase activation by transautophosphorylation.
Inhibition:
Maintained in an inactive, closed conformation by an interaction between the kinase domain and the negative autoregulatory C-terminal coiled-coil region.
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
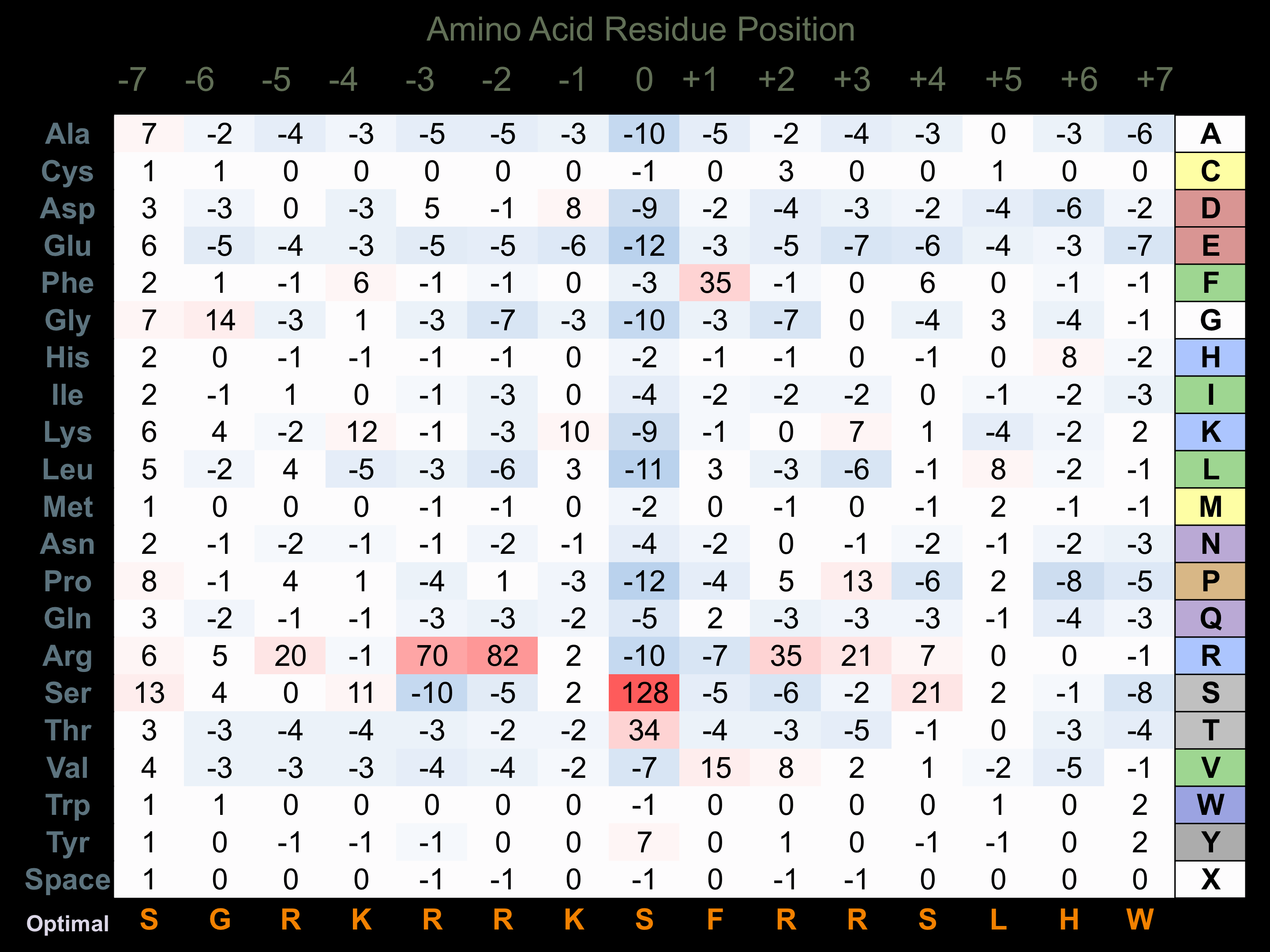
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | Kd = 37 nM | 5279 | 22037378 | |
| Foretinib | Kd = 180 nM | 42642645 | 1230609 | 22037378 |
| PD173955 | Kd = 250 nM | 447077 | 386051 | 22037378 |
| SB203580 | Kd = 400 nM | 176155 | 10 | 22037378 |
| A674563 | Kd = 460 nM | 11314340 | 379218 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Dasatinib | Kd = 1.2 µM | 11153014 | 1421 | 22037378 |
| Doramapimod | Kd = 1.2 µM | 156422 | 103667 | 22037378 |
| KW2449 | Kd = 2.1 µM | 11427553 | 1908397 | 22037378 |
| Vandetanib | Kd = 2.2 µM | 3081361 | 24828 | 22037378 |
| Lestaurtinib | Kd = 2.5 µM | 126565 | 22037378 | |
| Erlotinib | Kd = 3.4 µM | 176870 | 553 | 22037378 |
Disease Linkage
General Disease Association:
Musculoskeletal disorders
Specific Diseases (Non-cancerous):
Myotonic dystrophy
Comments:
Myotonic dystrophy is an inherited muscular disease, characterized by wasting of muscles (muscular dystrophy), heart defects, cataracts, changes in endocrine function, and myotonia. This disease is the most common adult-onset muscular dystrophy, typically beginning over the ages of 20-30 years. Myotonic dystrophy is inherited in an autosomal dominant manner.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 24447 diverse cancer specimens. This rate is only -22 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.28 % in 864 skin cancers tested; 0.28 % in 1229 large intestine cancers tested; 0.2 % in 589 stomach cancers tested; 0.14 % in 273 cervix cancers tested; 0.1 % in 1512 liver cancers tested; 0.09 % in 603 endometrium cancers tested; 0.09 % in 1608 lung cancers tested.
Frequency of Mutated Sites:
None > 1 in 20,204 cancer specimens
Comments:
Only 6 deletions, 1 complex mutation and no insertions noted on the COSMIC website.

