Nomenclature
Short Name:
DYRK1B
Full Name:
Dual-specificity tyrosine-phosphorylation regulated kinase 1B
Alias:
- Dual-specificity tyrosine- (Y)-phosphorylation regulated kinase 1B
- Dual-specificity tyrosine-phosphorylation regulated kinase 1B
- MIRK
- Mirk protein kinase
- DYR1B
- EC 2.7.12.1
- Kinase DYRK1B
- Minibrain-related kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
DYRK
SubFamily:
Dyrk1
Specific Links
Structure
Mol. Mass (Da):
69,198
# Amino Acids:
629
# mRNA Isoforms:
3
mRNA Isoforms:
69,198 Da (629 AA; Q9Y463); 66,336 Da (601 AA; Q9Y463-3); 64,934 Da (589 AA; Q9Y463-2)
4D Structure:
Dimer. Interacts with DCOHM, MAP2K3/MKK3, RANBP9 and TCF1/HNF1A. Part of a complex consisting of RANBP9, RAN, DYRK1B and COPS5. Interatcs with WDR68.
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 111 | 431 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K57 (N6).
Serine phosphorylated:
S276, S486.
Tyrosine phosphorylated:
Y63, Y64+, Y111, Y271+, Y273+, Y485.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 91
91
696
29
1166
 7
7
57
14
88
 19
19
144
33
386
 36
36
273
111
482
 83
83
639
24
512
 1
1
8
74
6
 31
31
234
31
519
 58
58
442
57
597
 42
42
319
17
190
 16
16
120
101
222
 14
14
106
50
271
 72
72
553
184
569
 17
17
133
55
374
 7
7
54
9
61
 22
22
172
42
392
 5
5
39
14
26
 10
10
75
137
187
 93
93
716
38
1974
 41
41
317
108
552
 36
36
278
109
325
 21
21
164
44
297
 23
23
174
46
324
 100
100
766
35
2139
 53
53
403
42
546
 26
26
196
40
388
 59
59
449
75
535
 17
17
131
58
268
 28
28
218
40
381
 22
22
166
41
320
 31
31
238
28
213
 72
72
552
24
542
 96
96
736
36
660
 8
8
62
67
154
 89
89
684
52
592
 24
24
187
35
518
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
100 99.5
99.5
99.6
99.5 -
-
-
99 -
-
-
79 90.2
90.2
90.5
99 -
-
-
- 97.3
97.3
97.9
97 54.7
54.7
64.4
97.5 -
-
-
- 52.7
52.7
61.7
- 32.1
32.1
46
- 55.4
55.4
65.3
79.5 64.7
64.7
72.2
77 -
-
-
- 45.1
45.1
54.1
- -
-
-
- -
-
-
50 -
-
-
- -
-
-
- -
-
-
- -
-
-
33 21.1
21.1
36
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | DCAF7 - P61962 |
| 2 | RANBP9 - Q96S59 |
| 3 | PCBD1 - P61457 |
| 4 | MAP2K3 - P46734 |
| 5 | PCBD2 - Q9H0N5 |
| 6 | HNF1A - P20823 |
| 7 | MAPK1 - P28482 |
| 8 | MBP - P02686 |
| 9 | MAPK14 - Q16539 |
| 10 | RPS27A - P62988 |
Regulation
Activation:
NA
Inhibition:
Inhibited by RANBP9.
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Cip1 (p21, CDKN1A) | P38936 | S152 | SMTDFYHSKRRLIFS | - |
| Cyclin D1 (CCND1) | P24385 | T286 | EEVDLACTPTDVRDV | |
| Cyclin D1 (CCND1) | P24385 | T288 | VDLACTPTDVRDVDI | |
| GYS1 | P13807 | S641 | YRYPRPASVPPSPSL | - |
| GYS2 | P54840 | S641 | FKYPRPSSVPPSPSG | - |
| H3.1 | P68431 | T45 | PHRYRPGTVALREIR | |
| HNF1A | P20823 | S249 | IQRGVSPSQAQGLGS | |
| p27Kip1 | P46527 | S10 | NVRVSNGSPSLERMD | - |
| SF3B1 | O75533 | T434 | PARKLTATPTPLGGM |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
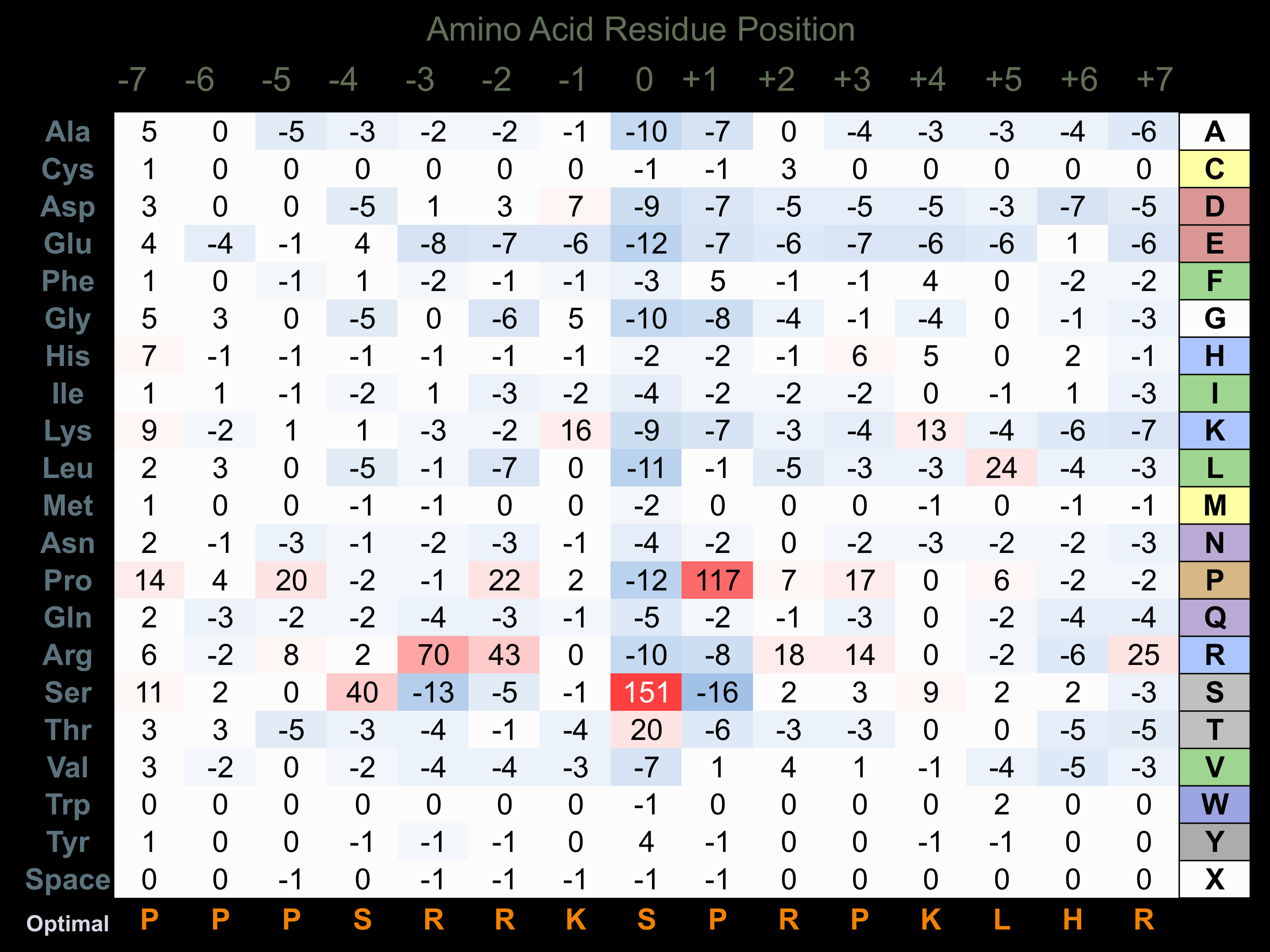
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Metabolic disorders
Specific Diseases (Non-cancerous):
Abdominal Obesity-Metabolic syndrome 3 (AOMS3)
Comments:
H90P and R102C mutations in DYRK1B link to Abdominal obesity-metabolic syndrome 3 (AOMS3), which is a disorder characterized by abdominal obesity, high triglycerides, low levels of high density lipoprotein cholesterol, high blood pressure, and elevated fasting glucose levels. AOMS3 is also a characteristic syndrome of early-onset coronary artery disease, central obesity, hypertension, and diabetes. It is mainly found in muscle and testis and is proposed to involve in nuclear function regulation. L28P variant of DYRK1B has been found to have a significant protection in 42 heterozygotes against type 2 diabetes and a trend to has a protective effect against hypertension.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for DYRK1B in diverse human cancers of 633, which is 1.4-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 68 for this protein kinase in human cancers was 1.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24847 diverse cancer specimens. This rate is only -7 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.51 % in 1093 large intestine cancers tested; 0.2 % in 805 skin cancers tested; 0.13 % in 1620 lung cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,010 cancer specimens
Comments:
Only 7 deletion, 2 insertions and no complex mutations are noted on the COSMIC website.

