Nomenclature
Short Name:
EPHA1
Full Name:
Ephrin type-A receptor 1
Alias:
- EC 2.7.10.1
- EPA1
- EPHT1
- MGC163163
- Tyrosine-protein kinase receptor EPH
- EPH
- EPH receptor A1
- Ephrin type-A receptor 1
- EPHT
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Eph
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
108,067
# Amino Acids:
976
# mRNA Isoforms:
3
mRNA Isoforms:
108,127 Da (976 AA; P21709); 52,618 Da (490 AA; P21709-2); 51,069 Da (474 AA; P21709-3)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
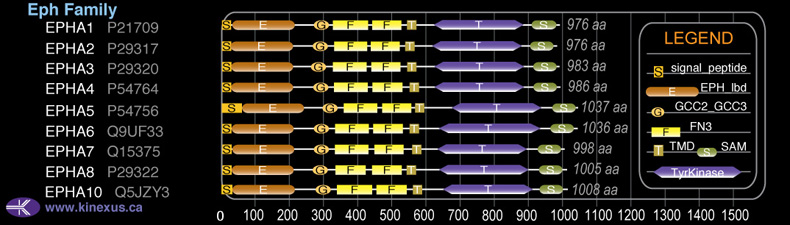
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N414.
Serine phosphorylated:
S34, S153, S811, S906, S908, S910, S919, T33, T155.
Threonine phosphorylated:
T780+, T783+, T805, T892.
Tyrosine phosphorylated:
Y599, Y605, Y781+.
Ubiquitinated:
K593, K597, K787.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 17
17
569
29
1330
 0.5
0.5
17
13
24
 4
4
128
15
219
 23
23
788
101
2144
 16
16
535
22
506
 4
4
119
86
274
 5
5
178
31
400
 28
28
937
45
2435
 13
13
454
17
460
 1
1
47
72
58
 3
3
108
39
141
 16
16
528
179
579
 3
3
98
42
221
 0.6
0.6
21
10
27
 3
3
101
31
200
 0.4
0.4
14
14
17
 1
1
34
128
91
 18
18
595
27
2664
 0.8
0.8
28
95
60
 12
12
414
109
498
 2
2
52
29
104
 1
1
47
31
86
 3
3
107
30
145
 2
2
80
29
209
 3
3
95
31
174
 24
24
809
68
1366
 3
3
89
45
116
 4
4
120
28
248
 2
2
58
27
109
 5
5
185
28
232
 26
26
888
24
846
 100
100
3379
36
7710
 5
5
179
63
482
 21
21
707
57
667
 4
4
136
35
82
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 37.6
37.6
54.4
99 37.6
37.6
54.4
97 -
-
-
88 -
-
-
- 96.8
96.8
98.5
90 86.8
86.8
91
- 88.3
88.3
92.5
88 44.3
44.3
61.1
88 44.3
44.3
61.1
- -
-
-
- 45.1
45.1
62.9
64 44.9
44.9
62.7
- 45.8
45.8
62.4
- 45.8
45.8
62.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
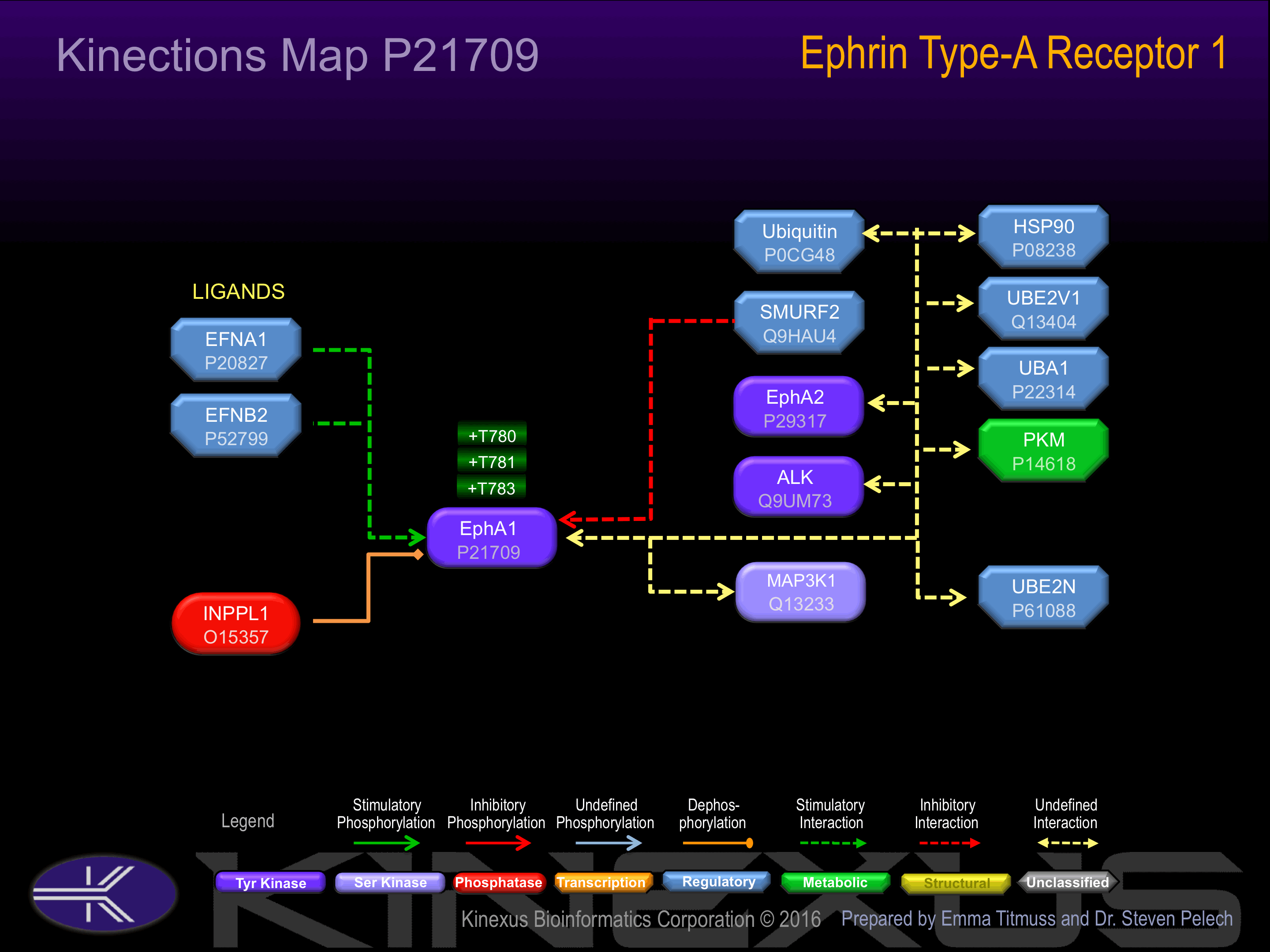
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | EFNA1 - P20827 |
| 2 | SMURF2 - Q9HAU4 |
Regulation
Activation:
Activated by binding of ephrin-A1 on a presenting cell, which is likely to induce dimerization and autophosphorylation.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
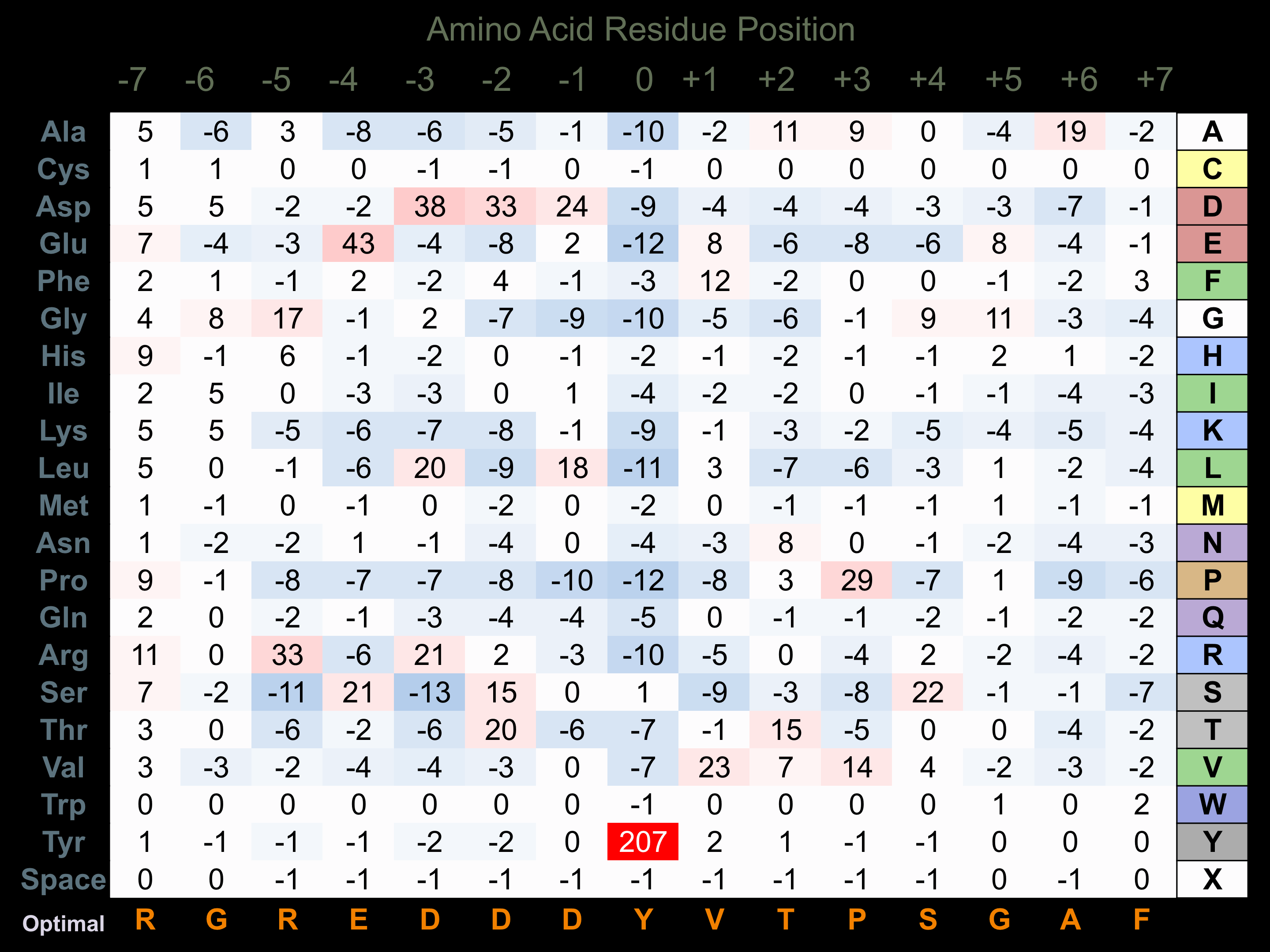
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Alzheimer's disease (AD)
Comments:
EphA1 is one of the common variants associated with late-onset Alzheimer's disease. Inhibition of the activation of EphA class receptors has been shown to inhibit angiogenesis in angiogenesis-dependet pancreatic islet cell carcinomas and the 4T1 model of metastatic mammary adenocarcinomas.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -59, p<0.09); Bladder carcinomas (%CFC= +104, p<0.021); Cervical cancer (%CFC= +343, p<0.002); and Skin melanomas - malignant (%CFC= -58, p<0.045).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24895 diverse cancer specimens. This rate is only 12 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 4 in 20,745 cancer specimens
Comments:
Only 4 deletions, no insertions, and 1 complex mutation are noted on the COSMIC website.