Nomenclature
Short Name:
EPHA2
Full Name:
Ephrin type-A receptor 2
Alias:
- EC 2.7.10.1
- ECK
- SEK2
- SEK-2
- Tyrosine-protein kinase receptor ECK
- EPH receptor A2
- Epithelial cell kinase
- Kinase EphA2
- MPK-5
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Eph
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
108266
# Amino Acids:
976
# mRNA Isoforms:
2
mRNA Isoforms:
108,266 Da (976 AA; P29317); 54,305 Da (497 AA; P29317-2)
4D Structure:
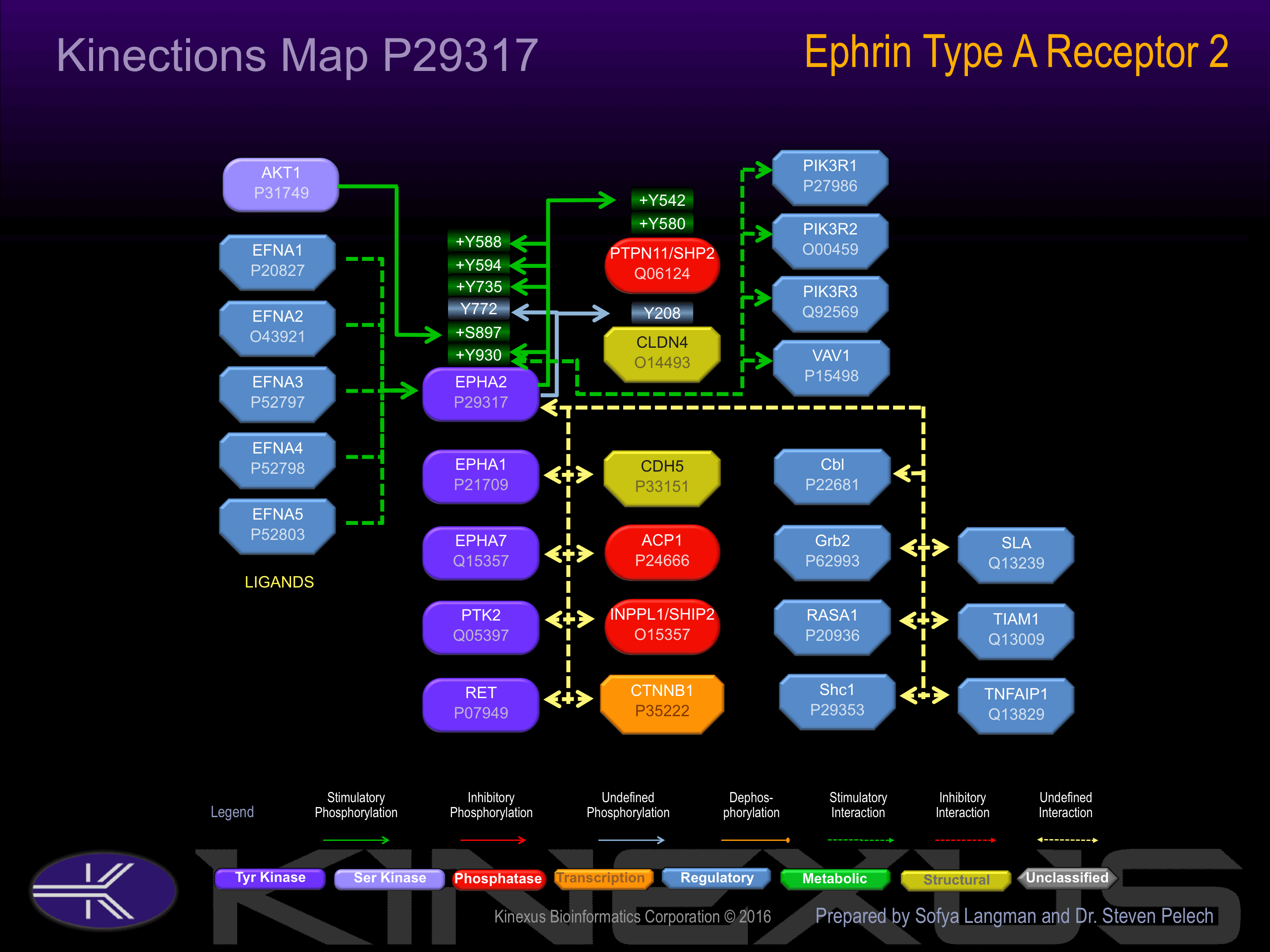
Monomer at low protein concentrations (around 10 µM) and forms homodimers at higher concentrations. Forms heterodimers with EFNA1. Interacts with SLA. The phosphorylated form interacts with VAV2, VAV3 and PI3-kinase p85 subunit By similarity. Interacts with INPPL1/SHIP2.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
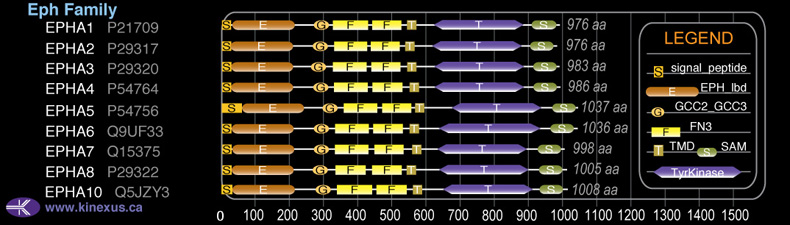
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Tyrosine phosphorylated:
Y67, Y575, Y588+, Y594+, Y628, Y685, Y694, Y735+, Y772, Y791, Y921, Y930+, Y960.
Ubiquitinated:
K136, K141, K162, K578, K583, K586, K617, K639, K655, K686, K778, K882, K945.
N-GlcNAcylated:
N407, N435.
Serine phosphorylated:
S113, S114, S153, S373, S426, S433, S570, S577, S579, S712, S749, S756, S761, S775, S790, S869, S880, S892, S897, S899, S901, S910, S961.
Threonine phosphorylated:
T437, T587, T593, T647, T692, T771, T773, T774, T898, T922, T940.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 32
32
824
16
992
 2
2
52
8
32
 9
9
217
17
305
 85
85
2164
71
4607
 29
29
733
14
587
 6
6
153
42
104
 12
12
312
19
534
 85
85
2146
48
3637
 14
14
347
10
315
 4
4
95
43
55
 5
5
123
30
98
 24
24
603
121
639
 6
6
142
28
110
 2
2
45
9
47
 6
6
148
25
176
 3
3
72
8
60
 12
12
297
117
1538
 12
12
298
24
403
 2
2
56
59
41
 25
25
641
56
658
 4
4
102
26
105
 4
4
112
28
110
 5
5
136
26
96
 5
5
136
23
251
 4
4
110
25
140
 100
100
2537
56
3559
 7
7
186
31
101
 12
12
303
24
199
 7
7
176
24
254
 2
2
44
14
22
 21
21
533
18
391
 37
37
929
21
1185
 9
9
226
49
515
 24
24
603
31
596
 40
40
1021
22
2477
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 38.9
38.9
57.8
98 97.8
97.8
98.3
98 -
-
-
93 -
-
-
- 94.8
94.8
96.9
95 -
-
-
- 92.6
92.6
95.3
93 50.6
50.6
66.1
93 -
-
-
- 84.8
84.8
90.8
- 52.2
52.2
68
76 52.5
52.5
68.7
74 50.6
50.6
68.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | EFNA2 - O43921 |
| 2 | TIAM1 - Q13009 |
| 3 | CBL - P22681 |
| 4 | ACP1 - P24666 |
| 5 | PIK3R2 - O00459 |
| 6 | TNFAIP1 - Q13829 |
| 7 | PIK3R1 - P27986 |
| 8 | EGFR - P00533 |
Regulation
Activation:
Activated by binding ephrin-A1, A2, A3, A4 or A5. Phosphorylation of Tyr-735 increases phosphotranserase activity and interaction with PIK3R1.
Inhibition:
NA
Synthesis:
By ultraviolet radiation (UV) via a TP53/p53-independent, but MAPK-dependent, mechanism
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
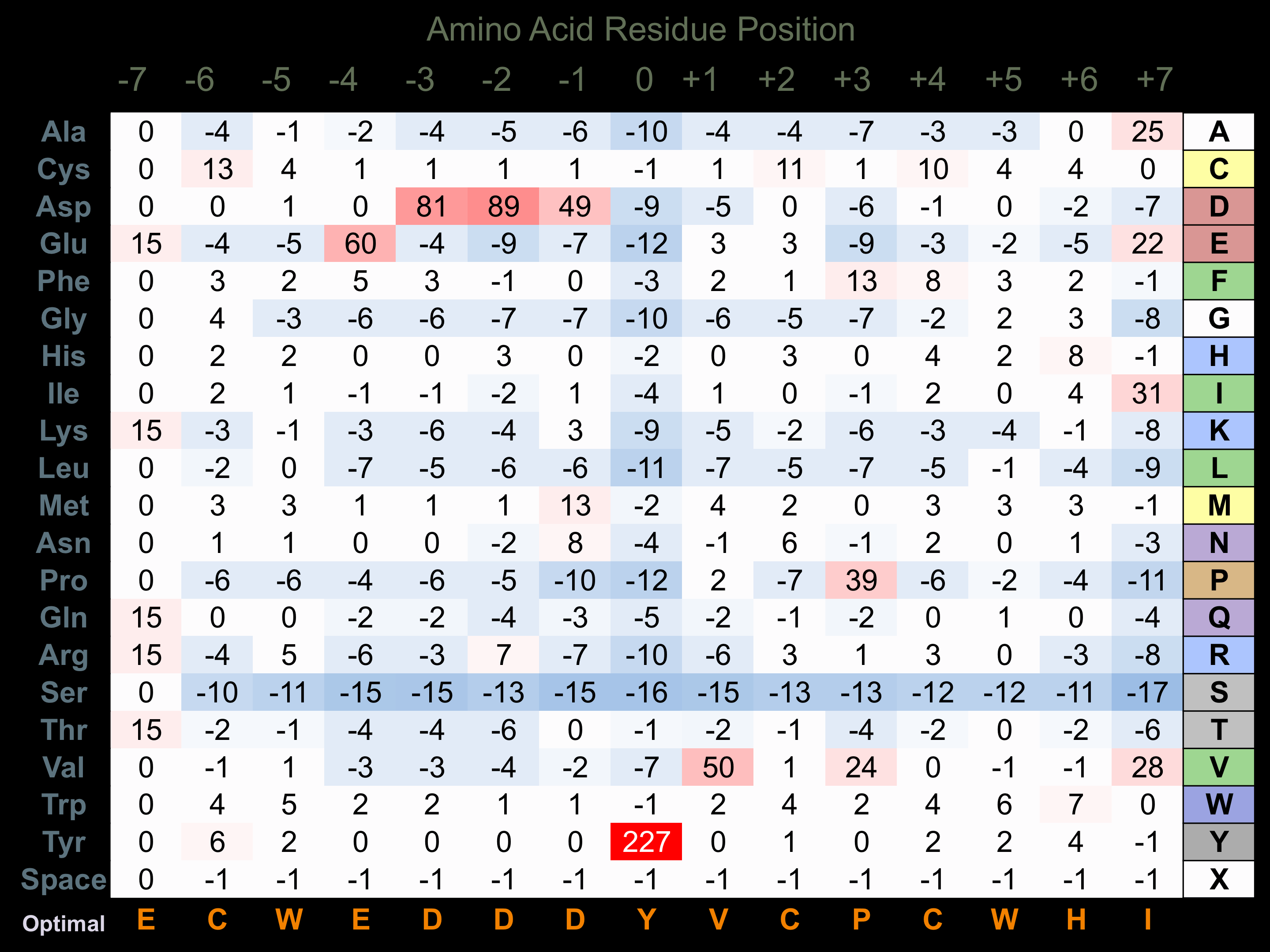
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
Matrix Type:
Experimentally derived from alignment of 5 known protein substrate phosphosites and 68 peptides phosphorylated by recombinant EphA2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, eye disorders
Specific Diseases (Non-cancerous):
Cataract; Cataract 6, multiple Types; Cataract 20, multiple Types; Cataract, age-related cortical, 2
Comments:
A cataract is an abnormal condition of the eyes characterized by the clouding of the lens, which can detrimentally affect visual acuity. It is predicted that ~50% of Americans over the age of 80 have cataracts and require surgical intervention for the symptomatic improvement. Symptoms of cataracts include blurry vision, faded colour vision, halo around lights, reduced night vision, and double vision. EphA2 is a receptor protein-tyrosine kinase that displays promiscuous binding to membrane-bound ligands of the ephrin-A family on adjacent cells. This signalling interaction regulates integrin-dependent adhesion, cellular migration, proliferation, and differentiation. EphA2 also participates in bone remodeling by directly regulating both osteoclastogenesis and osteoblastogenesis. Several mutations in the EphA2 gene have been observed in cataract patients, including substitution mutations (G948W, T940I, R721Q), a deletion mutation resulting in a frameshift (2915delTG), and splice site mutations. When expressed in a HEK293 cell culture, the R271Q mutant EphA2 protein displayed significantly elevated kinase catalytic activity compared to the wildtype protein, which resulted in a substantial reduction in ERK1/2 activity. Additionally, the R271Q mutant EphA2 protein displayed intracellular retention in certain organelles, which is not observed for wildtype proteins and potentially represents a pathological occurence. In animal studies, mice lacking the EphA2 gene displayed significant opacity of the lens at 3-4 months of development, followed by the formation of murine cataracts at 6-8 months, and eventual rupture of the lens. In addition, EphA2 expression was reported to decrease with age in wild-type mice, indicatingly a potential causal explanation for the predominance of cataracts in the aged population. In murine tissue, EphA2 expression in anterior epithelial cells is low initially, but upregulated as the cells enter differentiation, and is highly expressed in cortical fibre cells, but is absent in the nuclei. Significantly increased expression of Hsp25 (mouse homolog of human heat-shock protein 27, HSP27) has been observed in the lens tissue of EphA2 knockout mice, indicating the presence of excessive cellular stress, misfolded proteins, and potentially the activation of autophagy, which may contribute to the pathogenesis of cataracts.
Specific Cancer Types:
Brainstem glioma
Comments:
EPHA2 may be an oncoprotein (OP). Signficantly elevated EphA2 expression has been observed in many types of human cancer and appears to be involved in mediating communication between RAS-PI3K/AKT and RAS-MAPK intracellular signalling pathways, which are both known to promote tumorigenesis. Additionally, EphA2 is required for UV-radiation induced apoptosis. UV-radiation is a potent carcinogen responsible for the development of melanoma. However, further characterization of melanoma cell lines revealed either a pro-apoptotic or anti-apoptotic role for EphA2 dependent upon the cellular context. Additionally, studies performed in breast cancer cell lines revealed a link between EphA2 expression levels and changes in cytoskeletal morphology and the recruitment of disintegrin and metalloprotease-10 (MMP10) enzymes, thus suggesting a role for theEphA2 protein in the promotion of tumour invasion and metastasis.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -46, p<0.058); Bladder carcinomas (%CFC= +61, p<0.094); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -71, p<0.01); Cervical epithelial cancer (%CFC= +81, p<0.016); Cervical cancer (%CFC= +211, p<0.0002); Cervical cancer stage 1B (%CFC= +134, p<0.077); Cervical cancer stage 2A (%CFC= +139, p<0.055); Colon mucosal cell adenomas (%CFC= +49, p<0.004); Colorectal adenocarcinomas (early onset) (%CFC= +68, p<0.063); Oral squamous cell carcinomas (OSCC) (%CFC= -57, p<0.048); Ovary adenocarcinomas (%CFC= +50, p<0.094); Skin fibrosarcomas (%CFC= -55); Skin melanomas - malignant (%CFC= -63, p<0.0001); Skin squamous cell carcinomas (%CFC= +123, p<0.027); and Vulvar intraepithelial neoplasia (%CFC= +117, p<0.002). The COSMIC website notes an up-regulated expression score for EPHA2 in diverse human cancers of 325, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25597 diverse cancer specimens. This rate is only 20 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.39 % in 629 stomach cancers tested; 0.36 % in 1152 large intestine cancers tested; 0.33 % in 805 skin cancers tested; 0.16 % in 934 upper aerodigestive tract cancers tested; 0.12 % in 1942 lung cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,729 cancer specimens
Comments:
Nineteen deletions, no insertions or complex mutations are noted on the COSMIC website.