Nomenclature
Short Name:
ERK7
Full Name:
Mitogen-activated protein kinase 15
Alias:
- BI916334
- Mitogen-activated protein kinase 15
- MK15
- EC 2.7.11.24
- ERK8
- Extracellular signal regulated kinase 8
- Extracellular signal-regulated kinase 8
- MAPK15
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
MAPK
SubFamily:
Erk7
Structure
Mol. Mass (Da):
59,832
# Amino Acids:
544
# mRNA Isoforms:
3
mRNA Isoforms:
59,832 Da (544 AA; Q8TD08); 31,112 Da (277 AA; Q8TD08-3); 28,696 Da (254 AA; Q8TD08-2)
4D Structure:
Interacts with CSK/c-Src, ABL1, RET and TGFB1I1.
1D Structure:
Subfamily Alignment
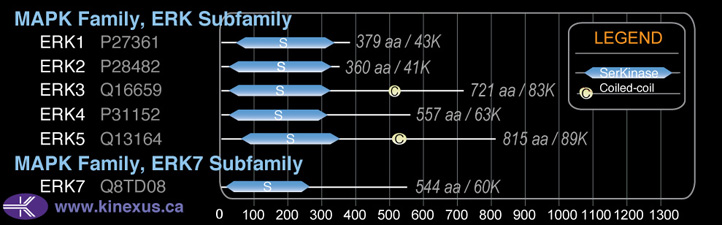
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 13 | 304 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K42.
Methylated:
R449.
Serine phosphorylated:
S192, S331, S362, S379, S415.
Threonine phosphorylated:
T175+, T352, T381.
Tyrosine phosphorylated:
Y177+Y340, .
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 56
56
1642
12
1506
 6
6
171
7
208
 -
-
-
-
-
 18
18
521
57
1766
 13
13
386
21
246
 0.3
0.3
9
18
8
 0.8
0.8
24
23
31
 8
8
230
7
265
 4
4
103
3
9
 3
3
80
55
125
 7
7
215
7
294
 29
29
845
19
494
 9
9
266
2
38
 16
16
453
5
752
 33
33
967
7
846
 1.5
1.5
43
12
85
 3
3
78
15
125
 6
6
187
5
137
 6
6
181
41
557
 13
13
376
61
207
 5
5
153
7
81
 3
3
74
5
16
 -
-
-
-
-
 16
16
471
7
566
 6
6
167
7
160
 25
25
736
39
963
 5
5
151
5
85
 17
17
486
5
581
 7
7
207
5
238
 6
6
183
28
184
 23
23
663
12
58
 100
100
2907
18
5167
 0.1
0.1
2
24
0
 24
24
707
52
608
 6
6
163
35
146
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
97 -
-
-
- -
-
-
72 -
-
-
- 71.5
71.5
77.8
73 -
-
-
- 69.2
69.2
75.5
72 68.7
68.7
75.1
72 -
-
-
- -
-
-
- 45.3
45.3
60
52 28.6
28.6
42
52.5 48.3
48.3
64.8
54 -
-
-
- -
-
-
48 34.6
34.6
50.5
- 42.6
42.6
61.2
59 44.9
44.9
62.6
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 29.7
29.7
42.8
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NR3C1 - P04150 |
| 2 | TGFB1I1 - O43294 |
| 3 | CSK - P41240 |
| 4 | SRC - P12931 |
| 5 | MBP - P02686 |
Regulation
Activation:
Activated by threonine and tyrosine phosphorylation.
Inhibition:
Inhibited by dual specificity phosphatases, such as DUSP1
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| ERK7 (MAPK15) | Q8TD08 | S192 | APEVLLSSHRYTLGV | |
| ERK7 (MAPK15) | Q8TD08 | S331 | AHEGVQLSVPEYRSR | |
| ERK7 (MAPK15) | Q8TD08 | S362 | EKGPEGVSPSQAHLH | |
| ERK7 (MAPK15) | Q8TD08 | S379 | RADPQLPSRTPVQGP | |
| ERK7 (MAPK15) | Q8TD08 | T175 | GPEDQAVTEYVATRW | + |
| ERK7 (MAPK15) | Q8TD08 | T352 | ECGGSSGTSREKGPE | |
| ERK7 (MAPK15) | Q8TD08 | T381 | DPQLPSRTPVQGPRP | |
| ERK7 (MAPK15) | Q8TD08 | Y177 | EDQAVTEYVATRWYR | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
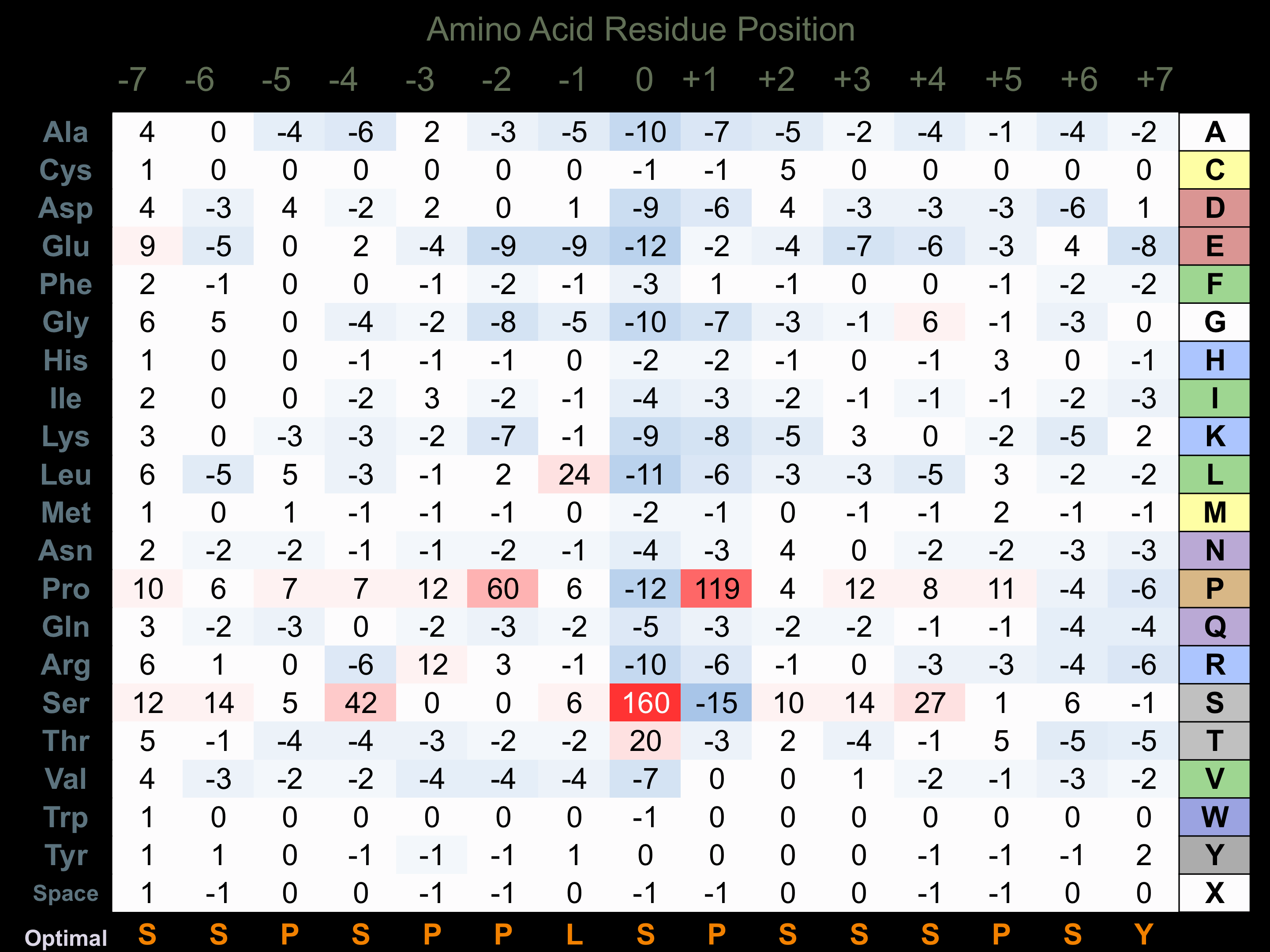
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for ERK7 in diverse human cancers of 661, which is 1.4-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Mutations at amino acid residues 42, 175, and 177 can lead to loss of ER7 autophosphorylation and phosphotransferase activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25346 diverse cancer specimens. This rate is only -11 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.47 % in 1093 large intestine cancers tested; 0.3 % in 805 skin cancers tested.
Frequency of Mutated Sites:
None > 2 in 19,689 cancer specimens
Comments:
Only 4 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

