Nomenclature
Short Name:
FGFR1
Full Name:
Basic fibroblast growth factor receptor 1
Alias:
- BFGFR
- FGFBR
- FGFR-1
- Fgr
- FLG; KAL2; Pfeiffer syndrome
- BFGF-R
- CD331
- CEK
- C-fgr
- EC 2.7.10.1
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
FGFR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
91,868
# Amino Acids:
822
# mRNA Isoforms:
21
mRNA Isoforms:
95,344 Da (853 AA; P11362-21); 91,868 Da (822 AA; P11362); 91,760 Da (822 AA; P11362-19); 91,668 Da (820 AA; P11362-4); 91,580 Da (820 AA; P11362-14); 90,618 Da (812 AA; P11362-20); 82,162 Da (733 AA; P11362-6); 81,962 Da (731 AA; P11362-8); 81,875 Da (731 AA; P11362-15); 74,133 Da (662 AA; P11362-10); 73,933 Da (660 AA; P11362-12); 73,475 Da (662 AA; P11362-2); 73,274 Da (660 AA; P11362-5); 63,769 Da (573 AA; P11362-7); 63,569 Da (571 AA; P11362-9); 55,740 Da (502 AA; P11362-11); 55,540 Da (500 AA; P11362-13); 33,412 Da (302 AA; P11362-17); 33,125 Da (300 AA; P11362-18); 16,487 Da (150 AA; P11362-16); 6,682 Da (61 AA; P11362-3)
4D Structure:
Interacts with SHB. Interacts with KLB By similarity. Interacts with KL and FGF23 By similarity. Interacts with GRB10.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
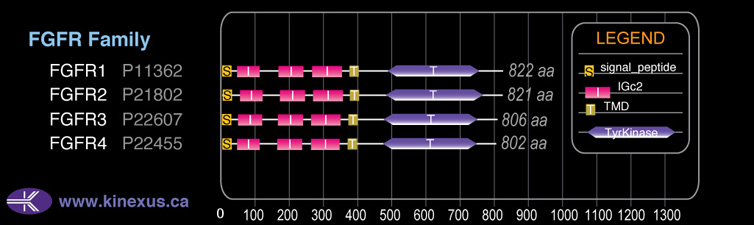
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S91, S410, S439, S447, S450, S451, S452, S588, S602, S777, S779, S789.
Threonine phosphorylated:
T428, T454.
Tyrosine phosphorylated:
Y154, Y210, Y280, Y307, Y463, Y572, Y583, Y585, Y605, Y613, Y653+, Y654+, Y677, Y701, Y730, Y766, Y776+.
Ubiquitinated:
K482, K638, K748.
Acetylated:
K164, K172.
N-GlcNAcylated:
N77, N117, N227, N240, N264,N296, N317, N330.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 73
73
1153
84
1103
 5
5
77
47
62
 48
48
761
88
594
 26
26
405
353
650
 58
58
912
85
721
 4
4
70
242
99
 17
17
273
107
557
 34
34
535
177
538
 25
25
398
51
316
 11
11
175
340
193
 6
6
98
147
109
 46
46
716
541
634
 12
12
192
154
188
 15
15
229
39
210
 45
45
709
134
736
 5
5
77
50
67
 13
13
210
562
154
 10
10
160
115
126
 9
9
145
340
110
 39
39
618
355
610
 23
23
362
129
336
 7
7
111
134
106
 13
13
200
102
166
 13
13
198
112
151
 12
12
189
127
154
 66
66
1040
239
2086
 11
11
166
163
166
 23
23
355
117
317
 30
30
465
117
414
 19
19
294
98
331
 11
11
166
48
199
 100
100
1571
100
4000
 48
48
749
190
1552
 51
51
802
187
704
 70
70
1095
109
2473
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.8
100 85.5
85.5
86.7
- -
-
-
99 -
-
-
100 98.9
98.9
99.4
99 -
-
-
- 98.4
98.4
99.2
98.5 97.7
97.7
98.7
98 -
-
-
- -
-
-
- 91.5
91.5
96.1
92 78.2
78.2
89.8
80 72.1
72.1
83.7
74 -
-
-
- 30.6
30.6
45.4
- -
-
-
- 30.7
30.7
47.1
- 35.6
35.6
53.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | FGF7 - P21781 |
| 2 | FGF3 - P11487 |
| 3 | KGFLP2 - Q2TVT3 |
| 4 | FGF6 - P10767 |
| 5 | FGF1 - P05230 |
| 6 | KGFLP1 - Q2TVT4 |
| 7 | FGF2 - P09038 |
| 8 | FRS2 - Q8WU20 |
| 9 | PLCG1 - P19174 |
| 10 | FRS3 - O43559 |
| 11 | NRP1 - O14786 |
| 12 | FGF5 - P12034 |
| 13 | MMP2 - P08253 |
| 14 | GRB14 - Q14449 |
| 15 | SOS1 - Q07889 |
Regulation
Activation:
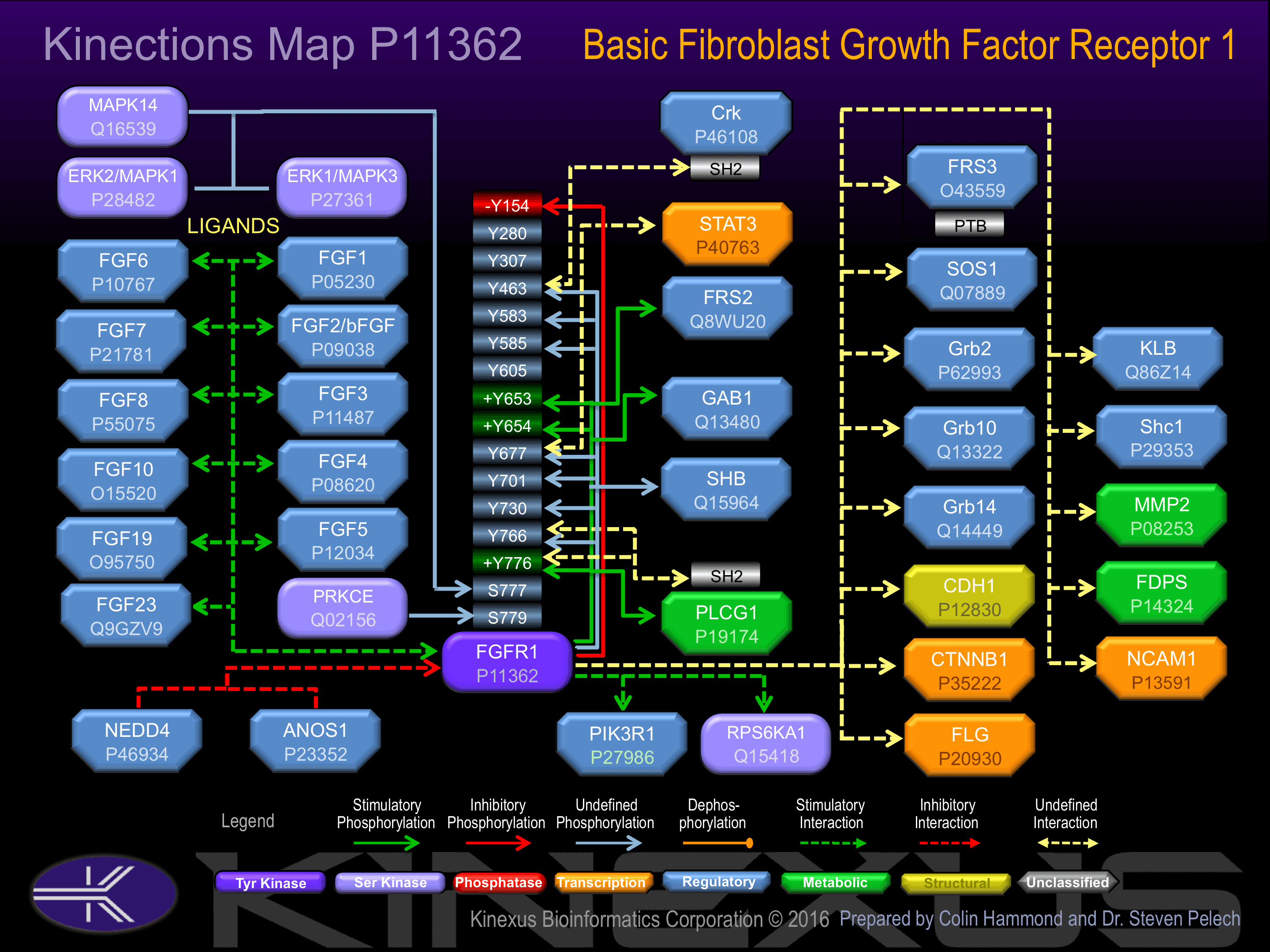
Activated by basic fibroblast growth factor (bFGF). Phosphorylation of Tyr-653 and Tyr-654 increases phosphotransferase activity. Phosphorylation of Tyr-766 induces interaction with PLCG1.
Inhibition:
Phosphorylation of Tyr-154 induces receptor internalization.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| FGFR1 | P11362 | Y154 | NRMPVAPYWTSPEKM | ? |
| FGFR1 | P11362 | Y463 | MLAGVSEYELPEDPR | ? |
| FGFR1 | P11362 | Y583 | RRPPGLEYCYNPSHN | |
| FGFR1 | P11362 | Y585 | PPGLEYCYNPSHNPE | |
| FGFR1 | P11362 | Y653 | RDIHHIDYYKKTTNG | + |
| FGFR1 | P11362 | Y654 | DIHHIDYYKKTTNGR | + |
| FGFR1 | P11362 | Y677 | EALFDRIYTHQSDVW | ? |
| FGFR1 | P11362 | Y701 | FTLGGSPYPGVPVEE | |
| FGFR1 | P11362 | Y730 | SNCTNELYMMMRDCW | ? |
| FGFR1 | P11362 | Y766 | ALTSNQEYLDLSMPL | |
| FGFR1 | P11362 | Y776 | FTLGGSPYPGVPVEE | + |
| p38a | Q16539 | S777 | SMPLDQYSPSFPDTR | ? |
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| FGFR1 | P11362 | Y154 | NRMPVAPYWTSPEKM | ? |
| FGFR1 | P11362 | Y463 | MLAGVSEYELPEDPR | ? |
| FGFR1 | P11362 | Y583 | RRPPGLEYCYNPSHN | |
| FGFR1 | P11362 | Y585 | PPGLEYCYNPSHNPE | |
| FGFR1 | P11362 | Y653 | RDIHHIDYYKKTTNG | + |
| FGFR1 | P11362 | Y654 | DIHHIDYYKKTTNGR | + |
| FGFR1 | P11362 | Y677 | EALFDRIYTHQSDVW | ? |
| FGFR1 | P11362 | Y701 | FTLGGSPYPGVPVEE | |
| FGFR1 | P11362 | Y730 | SNCTNELYMMMRDCW | ? |
| FGFR1 | P11362 | Y766 | ALTSNQEYLDLSMPL | |
| FGFR1 | P11362 | Y776 | FTLGGSPYPGVPVEE | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
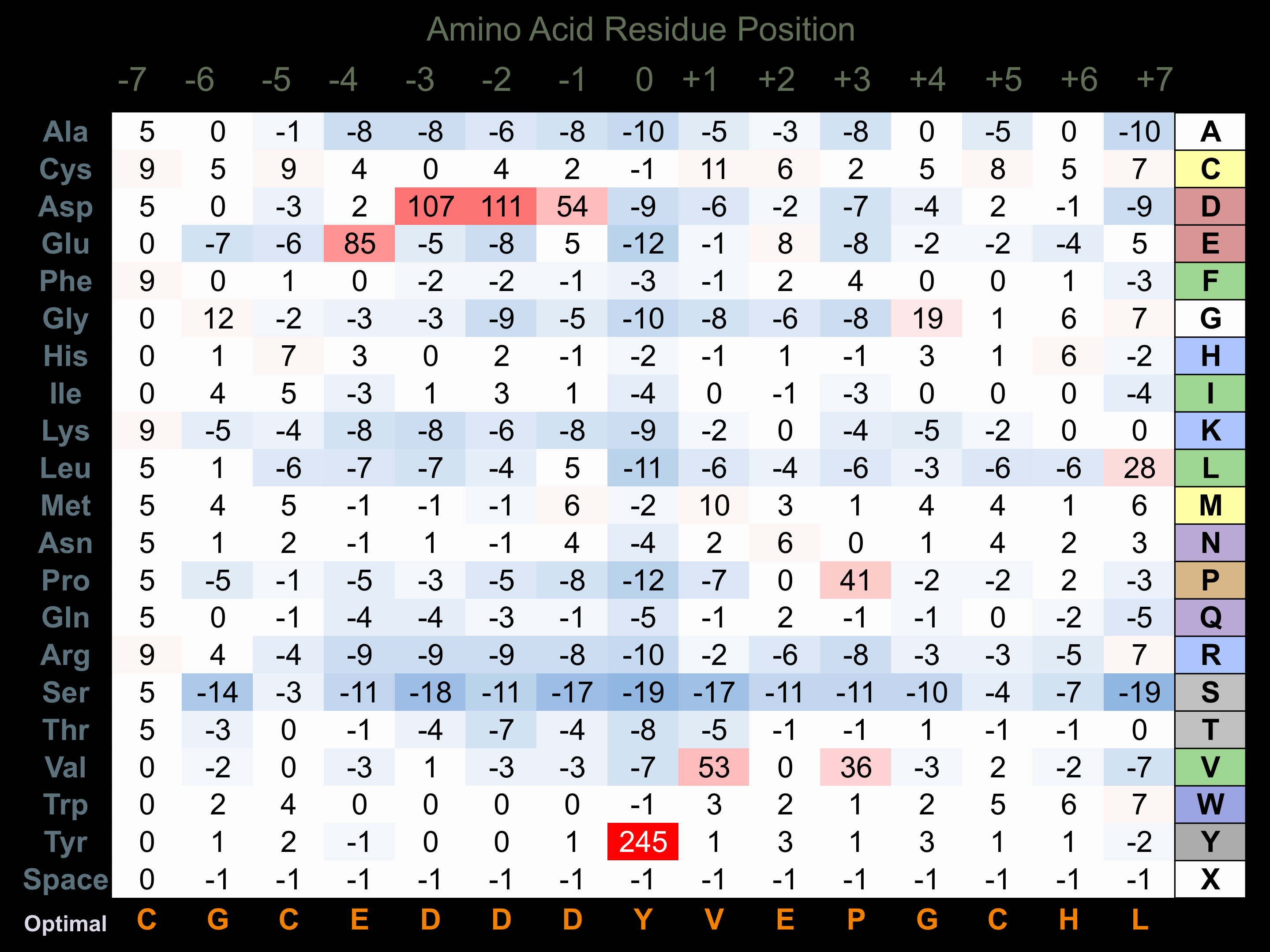
Matrix Type:
Experimentally derived from alignment of 17 known protein substrate phosphosites and 38 peptides phosphorylated by recombinant FGFR1 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, bone, development, and endocrine disorders
Specific Diseases (Non-cancerous):
Osteoglophonic dysplasia (OGD); Kallmann syndrome; Craniosynostosis (CSO); Hartsfield syndrome; Trigonocephaly 1 (HRTFDS); Cleft lip; Synostosis; Hypogonadotropic hypogonadism 2 with or without Anosmia (HH2); Crouzon syndrome (CFD1); Hypochondroplasia (HCH); Antley-Bixler syndrome; Infectious mononucleosis (ACS5); Septo-optic dysplasia (SOD); Kallmann syndrome 1; Achondroplasia (ACH); Saethre-Chotzen syndrome (SCS); Acrocephalosyndactylia (ACS1); Muenke syndrome; Plagiocephaly; Radioulnar synostosis; Pfeiffer syndrome Type 1 (PS); FGFR-related craniosynostosis syndromes; Kallmann syndrome 2; Hypogonadotropic Hypogonadism 8 with or without Anosmia; Hypogonadotropic hypogonadism 17 with or without Anosmia; Tooth agenesis, selective, 1, with or without orofacial cleft; FGFR1-related Craniosynostosis; Trigonocephaly, nonsyndromic; FGFR1-related isolated gonadotropin-releasing hormone deficiency; FGFR1-related isolated gonadotropin-releasing hormone deficiency
Comments:
Osteoglophonic Dysplasia (OGD) is characterized by abnormal bone growth, craniofacial deformations, and dwarfism. In OGD FGF-2 mediated activity and basal activity are increased with an FGFR1 mutation of Y374C. The rare disease Kallmann Syndrome can manifest with delayed puberty, impaired sense of smell, colour blind, cleft lip, hearing loss, kidney development issues, and infertility. Craniosynostosis (CSO) is a disorder arising from premature fusion of fibrous joints in the skull resulting in abnormal skull shape, yet often allowing full brain development. Sometimes many fusions occur in CSO, resulting in restricted brain growth leading to cognitive impairment and sometimes seizures and blindness. Hartsfield Syndrome (HRTFDS) is characterized by respiratory distress, and holoprosencephaly (facial malformation). HRTFDS can affect the olfactory bulb and lung tissues. Trigonocephaly 1 is a rare eye disease. Synostosis is rare disease resulting in the fusion of two bones, typically cranial bones. Hypogonadotropic Hypogonadism 2 with or Without Anosmia (HH2) is a disorder affecting the sexual organs, and is characterized by lack of sex hormone production. In HH2 a similar phenotype to normosmic idiopathic hypogonadotropic hypogonadism is observed with a G48S mutation. In HH2 incomplete glycosylation and reduced cell surface expression of FGFR1 can occur with the Y99C, Y228D, I239T mutations. In HH2 the A167S mutation can be associated with corpus callosum agenesis, cleft palate, unilateral deafness, and fusion of the fourth and fifth metacarpal bones. In HH2 severe ear defects occur with the C178S, and R622G mutations. In HH2 improper folding may occur with the G237S mutation. In HH2 receptor affinity for fibroblast growth factor is decreased with a R250Q mutation. In HH2 a phenotype similar to Kallmann syndrome occurs with a G348R mutation. Bimanual synkinesis occurs in HH2 with a V607M mutation. Tyrosine kinase activity in HH2 is impaired with the K618N, P722H, and N724K mutations. A patient suffering from HH2 had a cleft palate, iris coloboma, and unilateral absence of nasal cartilage had a mutation, P772S. Crouzon Syndrome is a rare disease arising from premature fusion of skull bones resulting in facial abnormalities. Hypochondroplasia (HCH) is a rare bone disease characterized by dwarfism. Antley-Bixler Syndrome is a rare bone disease resulting in malformations along the majority of the body. Infectious Mononucleosis (ACS5) inhibits the normal formation of the skull resulting in bulging, wide-set eyes, and an underdeveloped upper jaw. ACS5 can affect pharynx, bone, and spleen. Septo-Optic Dysplasia (SOD) is a rare genetic disorder which is characterized by abnormal development of the optic disk, absence of agenesis, defective pituitary gland, blindness, issues focusing eyes, and periodically intellectual disability. Achondroplasia (ACH) is a rare disease where conversion of cartilage to bone is inhibited. Saethre-Chotzen Syndrome (SCS) is a rare bone disease where skull bones fuse prematurely during development. Acrocephalosyndactylia (ACS1) is also a rare bone disease where skull bones fuse prematurely during development, and can affect bone, eye, or ovary tissues. Muenke Syndrome is a rare disease where the premature fusing of the coronal structure occurs, leading to facial abnormalities. Muenke Syndrome can also affect hands, feet, induce hearing loss, and result in developmental delay. Plagiocephaly is a rare disease where there is asymmetrical flattening of one side of a neonate's head. Radioulnar Synostosis is related to synostosis and craniosynostosis. Pfeiffer Syndrome Type 1 (PS) is a rare bone disease leading to proptosis (forward setting of the eye, past the eyelid), low set ears, and brachydactylyl (short fingers). A gain of function mutation in (PS) is P252R. Tooth Agenesis, Selective, 1, with or Without Orofacial Cleft is a rare disease and may or may not be characterized by an orofacial cleft, but most likely will have facial deformations, partial mandibula absence, and tooth shape anomaly. Trigonocephaly, Nonsyndromic a rare bone disease resulting in the premature fusion of the metopic structure (part of the forehead).
Specific Cancer Types:
Myeloid neoplasm associated with FGFR1 rearrangement; 8p11 myeloproliferative syndrome; Myeloproliferative disorder; Colorectal cancer; Gliomatosis cerebri; Osteochondromas; Lobular neoplasia; Pilocytic astrocytomas; Gliosarcomas; Giant cell glioblastomas
Comments:
FGFR1 may be an oncoprotein (OP). The active form of the protein kinase normally acts to promote tumour cell proliferation. Myeloid Neoplasm Associated with FGFR1 gene rearrangement and is a rare disease that typically affects myeloid tissues. Myeloid Neoplasm Associated with Fgfr1 Rearrangement is related to 8p11 myeloproliferative syndrome and chromosome 8p11 myeloproliferative syndrome. 8p11 Myeloproliferative Syndrome is a rare blood cancer forming either myeloid or lymphoid cell cancer. Myeloproliferative Disorder is a rare cancer disease affecting bone, bone marrow, and myeloid tissues. Myeloproliferative Disorder is related to Alzheimer’s and G protein signalling H-RAS regulation pathway. Gliomatosis Cerebri is a rare brain cancer resulting in a diffuse tumour that is therefore difficult to treat. Osteochondroma is a rare bone tumour. Lobular Neoplasia is a breast cancer, and is related to breast fibroadenoma. Pilocytic Astrocytoma is a benign tumour of the brain or spinal cord which is slow growing. Gliosarcoma is another rare cancer of the brain, but arising from glial cells. Giant Cell Glioblastoma is a multinucleated cell that has a large diameter.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -47, p<0.01); Brain glioblastomas (%CFC= -72, p<0.0001); Breast epithelial cell carcinomas (%CFC= +47, p<0.029); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +308, p<0.0006); Colorectal adenocarcinomas (early onset) (%CFC= +103, p<0.0001); Gastric cancer (%CFC= +69, p<0.033); Pituitary adenomas (aldosterone-secreting) (%CFC= +71, p<0.0002); Prostate cancer - primary (%CFC= -56, p<0.0001); Skin fibrosarcomas (%CFC= +268, p<0.007); Skin melanomas (%CFC= -53, p<0.032); Uterine fibroids (%CFC= +55, p<0.034); andVulvar intraepithelial neoplasia (%CFC= -47, p<0.001). The COSMIC website notes an up-regulated expression score for FGFR1 in diverse human cancers of 490, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. FGFR1 phosphotransferase activity can be abrogated with K514A and D623A mutations. Autophosphorylation and kinase activity can be inhibited with a mutation at both Y653F and Y654F. The first autophosphorylation can be sped up, but the second autophosphorylation event inhibited entirely with a N546K mutation. The R577E mutation can lead to strongly decreased autophosphorylation in response to FGF signalling. PLCG1 interaction with FGFR1 can be inhibited with mutation of R609V and D755V. The Y766F mutation can inhibit PLCG1 and SHB interaction with FGFR1, decrease FRS2 phosphorylation, and induce RAS or MAPK signalling, leading to cell proliferation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 30128 diverse cancer specimens. This rate is only 29 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.41 % in 1560 large intestine cancers tested; 0.31 % in 856 stomach cancers tested; 0.22 % in 1447 skin cancers tested; 0.19 % in 708 endometrium cancers tested; 0.19 % in 2189 central nervous system cancers tested; 0.09 % in 2253 lung cancers tested; 0.07 % in 2143 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: N546K (19); K656E (8); K656M (4).
Comments:
Only 1 deletion, 2 insertions and no complex mutations are noted on the COSMIC website.