Nomenclature
Short Name:
GCK
Full Name:
Mitogen-activated protein kinase kinase kinase kinase 2
Alias:
- B lymphocyte serine/threonine-protein kinase
- M4K2
- MAP4K2
- MAPK/ERK kinase kinase kinase 2
- RAB8IP
- BL44
- EC 2.7.11.1
- GCK
- Germinal center kinase
- Kinase GCK
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
KHS
Specific Links
Structure
Mol. Mass (Da):
91556
# Amino Acids:
820
# mRNA Isoforms:
2
mRNA Isoforms:
91,556 Da (820 AA; Q12851); 90,809 Da (812 AA; Q12851-2)
4D Structure:
Interacts with TRAF2, MAP3K1 and RAB8A.
1D Structure:
Subfamily Alignment
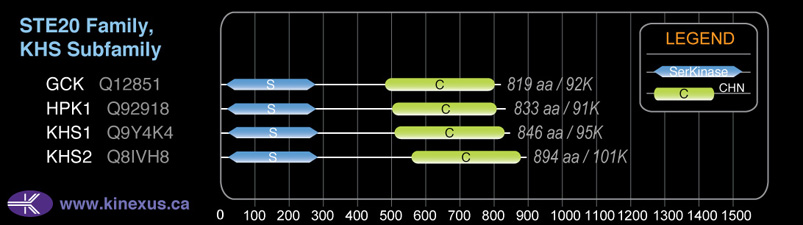
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S8+, S39, S164+, S170+, S227, S298, S328, S385, S394.
Threonine phosphorylated:
T174-, T306, T326, T574, T578, T819.
Tyrosine phosphorylated:
Y27, Y307.
Ubiquitinated:
K32.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 16
16
239
16
337
 4
4
63
10
81
 7
7
108
32
120
 22
22
321
81
721
 47
47
685
11
484
 0.2
0.2
3
37
4
 20
20
296
19
537
 49
49
727
53
1941
 21
21
313
10
244
 6
6
93
73
135
 5
5
72
43
88
 27
27
397
125
475
 10
10
146
43
223
 3
3
39
7
37
 8
8
120
41
152
 0.5
0.5
8
8
8
 3
3
40
133
54
 14
14
206
38
485
 2
2
31
74
44
 24
24
358
56
408
 7
7
96
41
113
 14
14
210
43
233
 5
5
73
41
77
 6
6
93
39
122
 13
13
192
41
212
 51
51
753
62
2425
 9
9
128
46
167
 6
6
83
39
91
 6
6
94
38
177
 1.4
1.4
21
14
5
 100
100
1473
18
1058
 30
30
446
21
514
 10
10
144
80
335
 42
42
621
31
617
 18
18
259
22
437
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
99.8
100 25.8
25.8
40.6
99.5 -
-
-
95 -
-
-
96 87.3
87.3
90.6
95 -
-
-
- 93.7
93.7
95.6
94 53.3
53.3
70
92 -
-
-
- 49.8
49.8
65.8
- 25.7
25.7
40
- 25.9
25.9
41
81 26.2
26.2
41.2
62 -
-
-
- 25.3
25.3
43.4
43 -
-
-
- 23.9
23.9
40.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
36 -
-
-
35 -
-
-
33 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | RAB8A - P61006 |
| 2 | MAP3K11 - Q16584 |
| 3 | TRAF2 - Q12933 |
| 4 | MAP2K4 - P45985 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
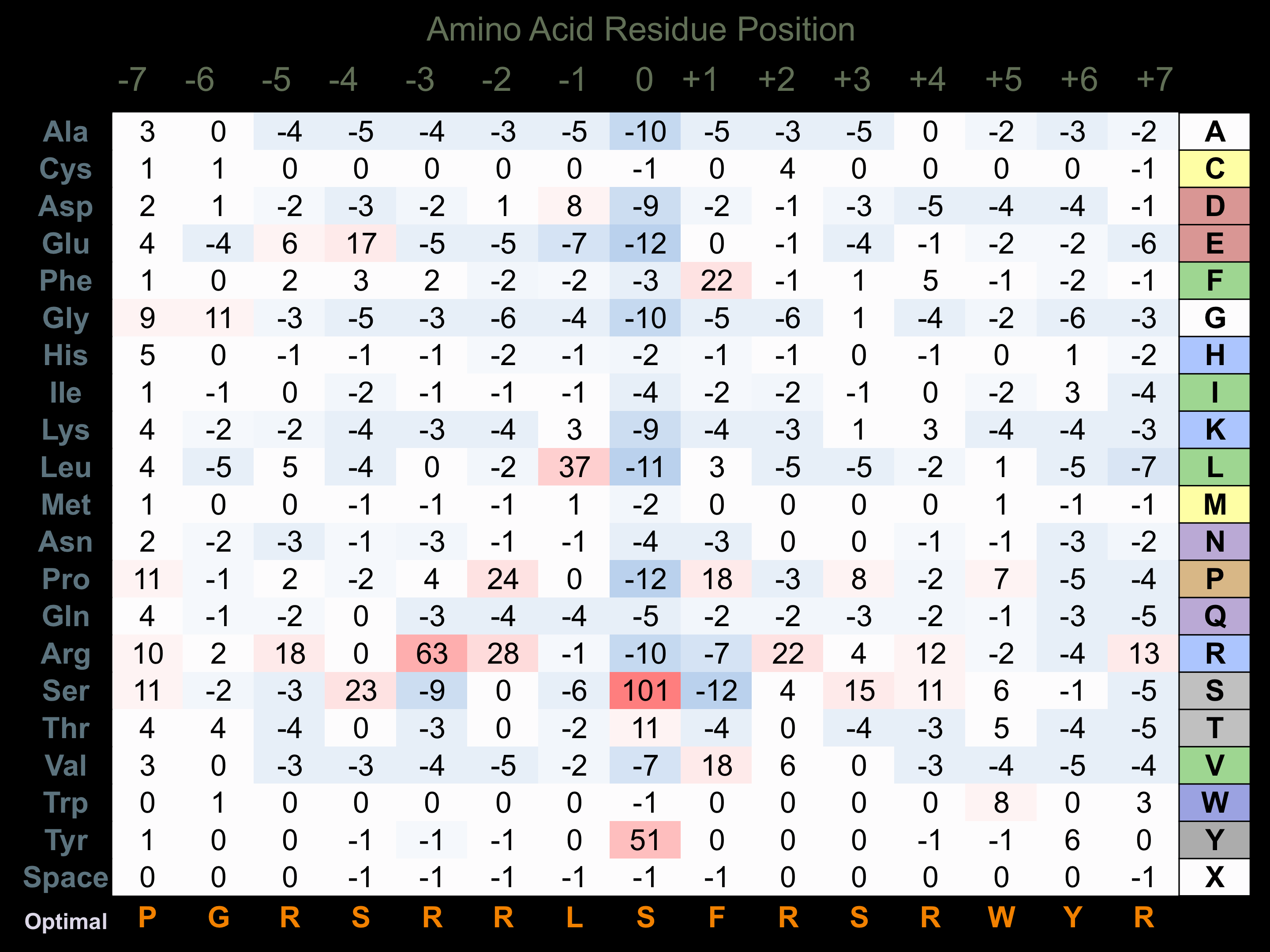
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, endocrine, and developmental disorders
Specific Diseases (Non-cancerous):
Maturity-onset diabetes of the young, Type 2 (MODY2); Gck-related hyperinsulinism; Gck-related permanent neonatal diabetes mellitus; Diabetes mellitus; Permanent neonatal diabetes mellitus (PNDM); Hypoglycemia; Hyperinsulinemic hypoglycemia; Maturity-onset diabetes of the young (MODY); Hyperglycemia; Hyperinsulinism; Gestational diabetes (GDM); Familial hyperinsulinism; Familial hyperinsulinism (FHI); Hyperinsulinemic hypoglycemia Familial 3; Monogenic diabetes; Pancreatic agenesis; 3-Hydroxyacyl-Coa dehydrogenase deficiency; Greig cephalopolysyndactyly syndrome (GCPS); Leucine-sensitive hypoglycemia of Infancy; Chromosome 7p deletion; Diabetes mellitus, Type 2; Maturity-onset diabetes of the young, Type 1; Diabetes mellitus, noninsulin-dependent, late onset
Comments:
Maturity-Onset Diabetes of the Young, Type 2 is a rare syndrome where insulin production is disrupted, leading to diabetes. Type 2 is an identifier for the maturity onset compared to type 1's juvenile onset. Gck-Related Hyperinsulinism is a rare disease resulting in Gck-mediated abnormally high levels of insulin in the blood. Gck-Related Permanent Neonatal Diabetes Mellitus is a rare disease characterised by abnormally high levels of blood sugar levels. Permanent Neonatal Diabetes Mellitus (PNDM) may be diagnosed through observation of slow growth before birth, epileptic episode, dehydration, hyperglycemia, and failure to thrive in infancy. The rare disease Hyperinsulinemic Hypoglycemia is characterized by high levels of insulin resulting in low blood glucose levels. Familial Hyperinsulinism is a rare condition characterized by high levels of insulin resulting in low blood glucose levels. The rare condition Hyperinsulinemic Hypoglycemia Familial 3 is related to hyperinsulinism due to glucokinase deficiency and to hyperinsulinemic hypoglycemia, familial, 1. Monogenic Diabetes differs from common Diabetes in that Monogenic Diabetes is characterized by a mutation in a single gene. The rare condition Pancreatic Agenesis is notable for the lack of development of a pancreas by the time of childbirth, leading to neonatal diabetes mellitus. 3-Hydroxyacyl-Coa Dehydrogenase Deficiency is characterized by the inability to convert fats to energy and can affect the liver, eye, and brain tissues. Greig Cephalopolysyndactyly Syndrome (GCPS) is a developmental condition where features can include polydactyly (extra toes or fingers), wide big toe or thumb (hallax), and fusion of fingers with a webbing tissue, widely spaced eyes, macrocephaly (large head), and more rarely seizures, intellectual impairment, or developmental delay. Leucine-Sensitive Hypoglycemia of Infancy is a rare condition related to hypoglycemia and permanent neonatal diabetes mellitus. The rare condition Chromosome 7p Deletion is related to Greig Cephalopolysyndactyly Syndrome (GCPS) where limb development is abnormal. Sufferers of Type 2 Diabetes Mellitus have insulin resistance and excess blood glucose levels. Maturity-Onset Diabetes of the Young, Type 1 is related to hypoglycemia and diabetes mellitus. Diabetes Mellitus, Noninsulin-Dependent, Late Onset is characterized by hyperglycemia (high blood glucose levels) induced by a reduced level of insulin production (but not fully abrogated, as would be the case in Type I diabetes).
Specific Cancer Types:
Insulinoma
Comments:
Insulinoma is a tumour of the pancreas arising from beta cells, and which will secrete insulin.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for GCK in diverse human cancers of 341, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24804 diverse cancer specimens. This rate is only -10 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R192Q (4).
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

