Nomenclature
Short Name:
GRK4
Full Name:
G protein-coupled receptor kinase 4
Alias:
- EC 2.7.11.16
- GPRK2L
- GPRK4
- ITI1
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
GRK
SubFamily:
GRK
Structure
Mol. Mass (Da):
66583
# Amino Acids:
578
# mRNA Isoforms:
4
mRNA Isoforms:
66,583 Da (578 AA; P32298); 63,030 Da (546 AA; P32298-2); 61,216 Da (532 AA; P32298-4); 57,663 Da (500 AA; P32298-3)
4D Structure:
Interacts with DRD3
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
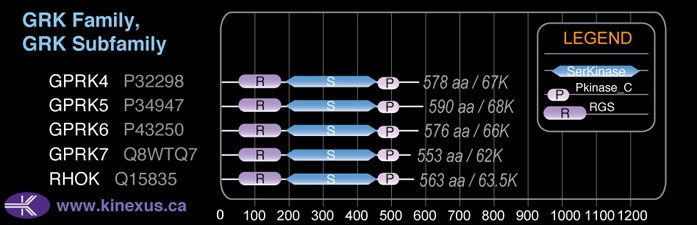
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
M1, K195, K527.
Palmitoylated:
True (site unclear, but possibly C563 by similarity with GRK6).
Serine phosphorylated:
S24, S485.
Ubiquitinated:
K216, K217.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 79
79
790
32
935
 1.3
1.3
13
14
12
 2
2
17
2
21
 18
18
179
108
334
 37
37
368
29
226
 15
15
148
90
519
 11
11
107
41
314
 83
83
830
31
1571
 27
27
272
17
234
 4
4
43
96
88
 3
3
26
21
25
 49
49
493
159
487
 0.8
0.8
8
24
9
 2
2
20
11
22
 3
3
32
15
26
 1.2
1.2
12
20
15
 7
7
69
173
556
 2
2
19
9
14
 2
2
20
95
23
 28
28
282
137
282
 5
5
55
15
61
 3
3
28
18
41
 0.9
0.9
9
12
5
 27
27
271
11
184
 5
5
48
14
93
 84
84
841
65
1550
 0.5
0.5
5
27
6
 1.2
1.2
12
10
18
 2
2
17
9
23
 7
7
66
42
86
 25
25
255
24
255
 51
51
510
41
800
 0.3
0.3
3
46
1
 60
60
597
83
554
 100
100
1001
48
1343
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.1
99.1
99.7
99 65.6
65.6
78.2
- -
-
-
78 -
-
-
- 76.5
76.5
84.6
79 -
-
-
- 75.3
75.3
86.3
76 75.4
75.4
87
76 -
-
-
- -
-
-
- 74.2
74.2
85.1
76 75.4
75.4
86.3
77 73.9
73.9
84.3
76 -
-
-
- 49
49
60.6
- -
-
-
- 50.8
50.8
66
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
Inhibited by heparin.
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
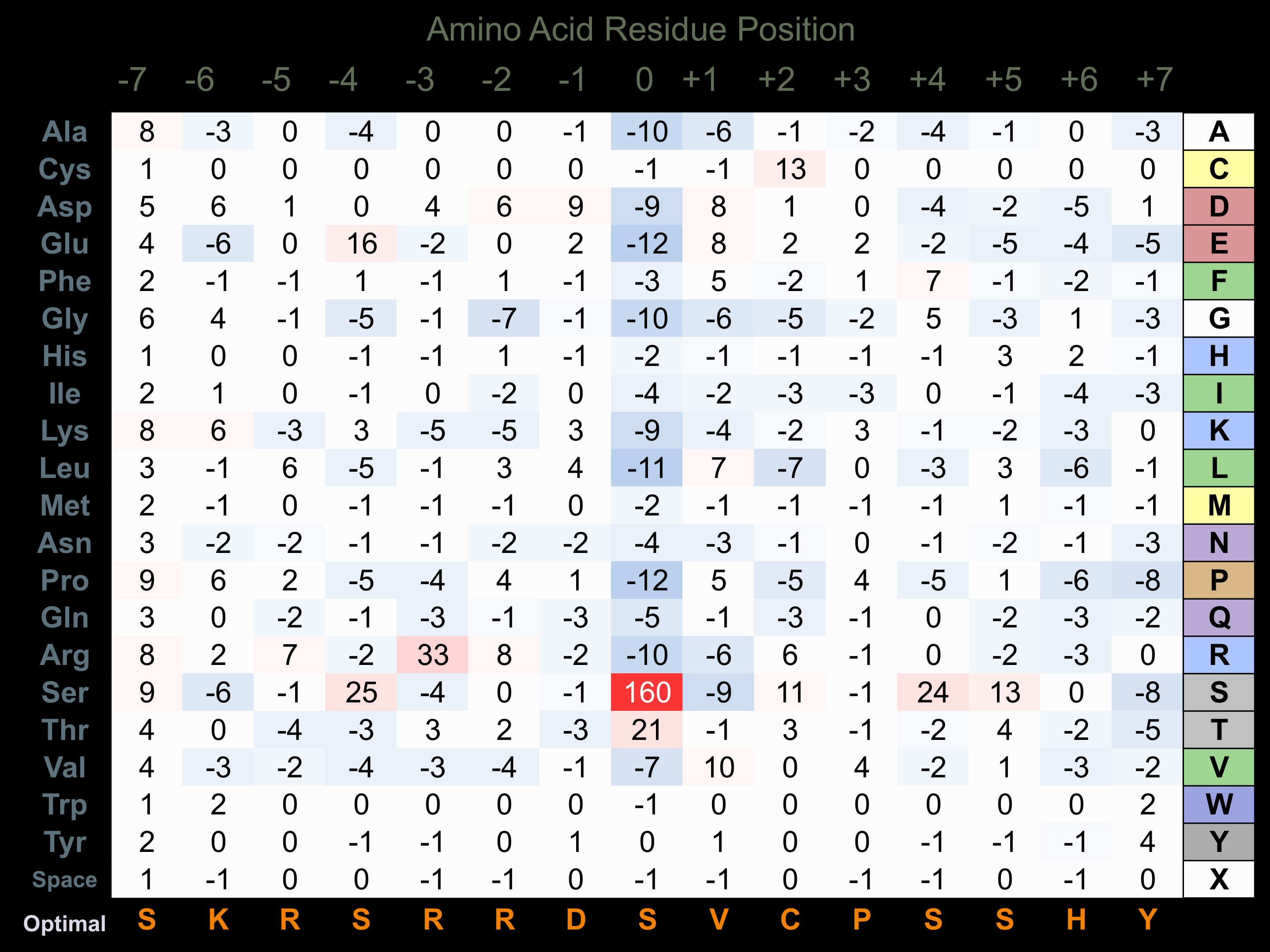
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular disorders
Specific Diseases (Non-cancerous):
Hypertension; Essential hypertension; Aortic coarctation
Comments:
Hypertension is a cardiovasculature disease characterized by high blood pressure. Essential hypertension, also called idiopathic hypertension, is hypertension that does not have an identifiable cause, which accounts for ~95% of all reported cases. Aortic coarctation, also called aortic narrowing, is a congenital cardiovascular disease characterized by the constriction of the aorta in the area where the ductus arteriosis inserts. As a result of this narrowing, the left ventricle must generate higher pressure to achieve the same flow of blood out of the heart, thus causing stress to the heart muscles. In extreme cases, the heart cannot push enough blood through the narrowing resulting in the lack of blood delivery to the lower half of the body. Mutations in GRK4 have been linked to hypertension. In general, GRKs are associated with both genetic and acquired hypertension as they function to desensitize G protein-coupled receptors, including D1 receptors (dopamine receptors). D1 receptors that are phosphorylated at basal levels on serine residues of the protein are expressed at higher levels in the renal proximal tubules in rodent models of hypertension and in human patients with hypertension, indicating a role for dopamine signalling in the development of hypertension. Additionally, single nucleotide polymorphisms (SNPs) in the GRK4-gamma subunit were observed in human patients with essential hypertension which were associated with increased GRK4 catalytic activity, resulting in elevated levels of serine phosphorylation of the D1 protein and the uncoupling of the D1 receptor from its G protein effector complex in the renal proximal tubule.
Comments:
GRK4 appears to be a tumour requiring protein (TRP), since it undergoes much fewer mutations than the typical protein in cancer cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Prostate cancer - primary (%CFC= +65, p<0.009). The COSMIC website notes an up-regulated expression score for GRK4 in diverse human cancers of 366, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 50 for this protein kinase in human cancers was 0.8-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0 % in 24900 diverse cancer specimens. This rate is -98 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a very low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.02 % in 942 upper aerodigestive tract cancers tested; 0.01 % in 1822 lung cancers tested.
Frequency of Mutated Sites:
None > 1 in 20,183 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

