Nomenclature
Short Name:
HCK
Full Name:
Tyrosine-protein kinase Hck
Alias:
- B-cell,myeloid kinase
- Kinase Hck
- P56-HCK and P60-HCK
- P59-Hck
- P60-Hck
- B-cell/myeloid kinase
- BMK
- EC 2.7.10.2
- Hemopoietic cell kinase
- JTK9
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Src
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
59600
# Amino Acids:
526
# mRNA Isoforms:
4
mRNA Isoforms:
59,600 Da (526 AA; P08631); 59,529 Da (525 AA; P08631-4); 57,312 Da (505 AA; P08631-2); 57,241 Da (504 AA; P08631-3)
4D Structure:
May interact (via SH3 domain) with HIV-1 Nef and Vif. This interaction would stimulates its tyrosine-kinase activity. Interacts (via SH3 domain) with HEV ORF3 protein. Interacts with ADAM15
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
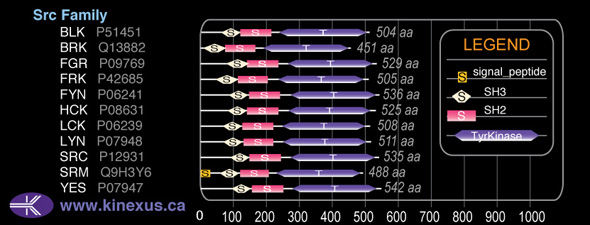
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K228.
Myristoylated:
G2.
Serine phosphorylated:
S119, S238, S340, S442, S462, S520.
Threonine phosphorylated:
T36, T202, T311, T412.
Tyrosine phosphorylated:
Y51, Y89, Y127, Y180, Y209, Y330, Y411+, Y522-.
Ubiquitinated:
K28, K124, K283, K286.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 27
27
1073
16
1438
 2
2
63
11
29
 2
2
85
13
75
 5
5
190
61
332
 17
17
653
14
646
 5
5
194
44
423
 13
13
503
23
621
 12
12
471
37
911
 14
14
566
10
423
 3
3
107
69
133
 2
2
76
30
56
 16
16
632
129
634
 4
4
161
24
66
 1.3
1.3
53
12
31
 2
2
74
25
100
 2
2
59
9
31
 0.7
0.7
29
198
30
 2
2
70
22
68
 1.2
1.2
47
61
34
 13
13
525
56
551
 3
3
107
26
56
 11
11
427
28
245
 2
2
67
22
75
 0.8
0.8
33
20
34
 3
3
117
26
83
 21
21
809
42
1603
 5
5
185
27
85
 3
3
99
22
157
 1.4
1.4
56
22
43
 3
3
116
14
80
 17
17
679
18
454
 13
13
503
21
607
 100
100
3930
17
1065
 49
49
1921
31
676
 2
2
72
22
45
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 80.3
80.3
81.1
97 61.1
61.1
72.9
98 -
-
-
92 -
-
-
92 76.7
76.7
79.9
92 -
-
-
- 89.7
89.7
94.9
90 89.5
89.5
94.5
90 -
-
-
- 68.8
68.8
82.3
- 64.1
64.1
78.3
79 56.6
56.6
72.4
68 42
42
47.3
77 -
-
-
- 52.3
52.3
67.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | BCR - P11274 |
| 2 | CBL - P22681 |
| 3 | RAPGEF1 - Q13905 |
| 4 | ABL1 - P00519 |
| 5 | PLAUR - Q03405 |
| 6 | WAS - P42768 |
| 7 | ADAM15 - Q13444 |
| 8 | CSF2RB - P32927 |
| 9 | VAV1 - P15498 |
| 10 | IL6ST - P40189 |
| 11 | FCGR1A - P12314 |
| 12 | ABL2 - P42684 |
| 13 | CCR3 - P51677 |
| 14 | CSK - P41240 |
| 15 | CSF3R - Q99062 |
Regulation
Activation:
NA
Inhibition:
Phosphorylation of Tyr-522 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Abl1 | P00519 | Y115 | QGWVPSNYITPVNSL | |
| Abl1 | P00519 | Y128 | SLEKHSWYHGPVSRN | |
| Abl1 | P00519 | Y139 | VSRNAAEYLLSSGIN | + |
| Abl1 | P00519 | Y172 | LRYEGRVYHYRINTA | |
| Abl1 | P00519 | Y185 | TASDGKLYVSSESRF | |
| Abl1 | P00519 | Y215 | GLITTLHYPAPKRNK | |
| Abl1 | P00519 | Y226 | KRNKPTVYGVSPNYD | + |
| Abl1 | P00519 | Y393 | RLMTGDTYTAHAGAK | + |
| Abl1 | P00519 | Y70 | PNLFVALYDFVASGD | + |
| Abl1 iso2 | P00519-2 | Y134 | QGWVPSNYITPVNSL | |
| Abl1 iso2 | P00519-2 | Y147 | SLEKHSWYHGPVSRN | |
| Abl1 iso2 | P00519-2 | Y158 | VSRNAAEYLLSSGIN | + |
| Abl1 iso2 | P00519-2 | Y191 | LRYEGRVYHYRINTA | |
| Abl1 iso2 | P00519-2 | Y204 | TASDGKLYVSSESRF | |
| Abl1 iso2 | P00519-2 | Y234 | GLITTLHYPAPKRNK | |
| Abl1 iso2 | P00519-2 | Y245 | KRNKPTVYGVSPNYD | + |
| Abl1 iso2 | P00519-2 | Y412 | RLMTGDTYTAHAGAK | + |
| Abl1 iso2 | P00519-2 | Y89 | PNLFVALYDFVASGD | + |
| ADAM15 | Q13444 | Y715 | LVMLGASYWYRARLH | |
| Bcr | P11274 | Y177 | ADAEKPFYVNVEFHH | + |
| ELMO1 | Q92556 | Y18 | AIEWPGAYPKLMEID | |
| ELMO1 | Q92556 | Y216 | VLNSHDLYQKVAQEI | |
| ELMO1 | Q92556 | Y395 | AKHHQDAYIRIVLEN | |
| ELMO1 | Q92556 | Y511 | SKLQNLSYTEILKIR | |
| ELMO1 | Q92556 | Y720 | IPKEPSNYDFVYDCN | |
| Hck | P08631 | Y411 | RVIEDNEYTAREGAK | + |
| Hck | P08631 | Y51 | ASPHCPVYVPDPTST | |
| Hck | P08631 | Y522 | YTATESQYQQQP___ | - |
| RapGEF1 | Q13905 | Y504 | APIPSVPYAPFAAIL | |
| STAT5B | P51692 | Y699 | TAKAVDGYVKPQIKQ | + |
| WASP | P42768 | Y291 | AETSKLIYDFIEDQG | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
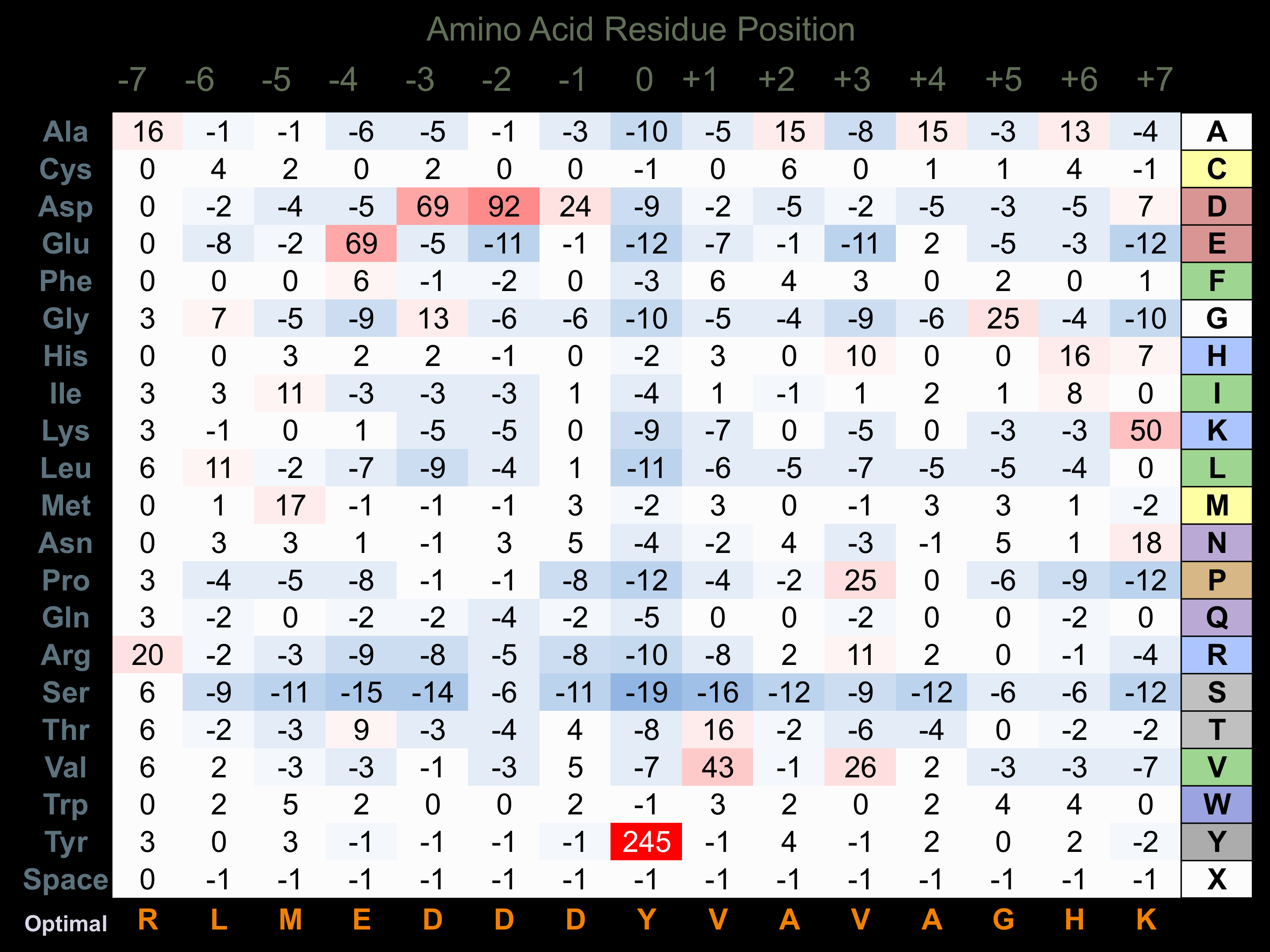
Matrix Type:
Experimentally derived from alignment of 28 known protein substrate phosphosites and 36 peptides phosphorylated by recombinant Hck in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Adenosquamous carcinomas; Cervical adenosquamous carcinomas
Comments:
HCK may be an oncoprotein (OP) based on its similarity to other Src family kinases that are known oncoproteins. The active form of the protein kinase normally acts to promote tumour cell proliferation. Hck has been linked with Adenosquamous carcinoma, which is a mixed lineage carcinoma made of squamous and gland cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Classical Hodgkin lymphomas (%CFC= +46, p<0.0001); Clear cell renal cell carcinomas (cRCC) (%CFC= +187, p<0.001); Large B-cell lymphomas (%CFC= +70, p<0.003); and Oral squamous cell carcinomas (OSCC) (%CFC= +110, p<0.001). The COSMIC website notes an up-regulated expression score for HCK in diverse human cancers of 326, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. A G3C mutation in HCK isoform 1 induces a moderate amount of palmitoylation, partially localizing the protein to the caveolar fraction. The G3S mutation completely inhibits palmitoylation, and prevents any cell membrane localization. The G23A mutation can inhibit both palmitoylation and myristoylation of Hck isoform 2. A C24S mutation can fully inhibit palmitoylation, and calveolar localizations in isoform 2 of Hck. Kinase activity can be fully lost with any of the K290E, E305A, or D381E mutations. Phosphotransferase activity can be impaired, yet affinity for target peptides increased with a Y411A mutation. The Y522F mutation can constitutively activate the kinase, leading to cellular perturbations and transformation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.13 % in 25186 diverse cancer specimens. This rate is 1.8-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.87 % in 805 skin cancers tested; 0.56 % in 1093 large intestine cancers tested; 0.36 % in 589 stomach cancers tested; 0.25 % in 1807 lung cancers tested; 0.1 % in 1962 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: P477L (4).
Comments:
Only 3 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

