Nomenclature
Short Name:
HH498
Full Name:
Serine-threonine-protein kinase TNNI3K
Alias:
- TNNI3-interacting kinase
- Cardiac ankyrin repeat kinase
- CARK
- TNNI3K
- LOC51086
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
MLK
SubFamily:
HH498
Structure
Mol. Mass (Da):
104,179
# Amino Acids:
936
# mRNA Isoforms:
4
mRNA Isoforms:
104,179 Da (936 AA; Q59H18-1); 94,517 Da (843 AA; Q59H18-4); 92,851 Da (835 AA; Q59H18); 77,957 Da (697 AA; Q59H18-3)
4D Structure:
Interacts with TNNI3, ACTC, ACTA1, MYBPC3, AIP, BABP3 and HADHB.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S424, S427, S814.
Threonine phosphorylated:
T581, T805.
Tyrosine phosphorylated:
Y62, Y416, Y425, Y771, Y788, Y804, Y810, Y812+.
Ubiquitinated:
K40.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 15
15
632
16
1213
 0.8
0.8
34
10
42
 0.9
0.9
37
25
31
 5
5
224
75
487
 6
6
254
12
218
 100
100
4194
53
10051
 7
7
314
19
620
 18
18
749
51
780
 5
5
195
10
201
 1
1
52
73
49
 0.7
0.7
29
36
35
 9
9
383
112
483
 0.5
0.5
20
36
23
 0.7
0.7
30
9
37
 0.9
0.9
37
35
54
 0.5
0.5
23
8
35
 1
1
43
186
32
 0.6
0.6
26
32
36
 0.7
0.7
30
66
26
 5
5
216
56
255
 1
1
46
38
28
 0.8
0.8
34
39
44
 0.4
0.4
17
25
25
 1
1
40
31
35
 0.8
0.8
35
38
45
 5
5
198
55
383
 0.4
0.4
17
39
21
 0.5
0.5
20
31
18
 0.6
0.6
24
31
29
 1
1
55
14
52
 9
9
386
18
279
 47
47
1951
21
3539
 4
4
149
47
399
 17
17
702
26
614
 7
7
273
22
265
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.2
98.2
98.3
99.5 96.5
96.5
97.2
98 -
-
-
96.5 -
-
-
- 80.1
80.1
82.6
95 -
-
-
- 80.7
80.7
84.4
91 81.3
81.3
85.3
91 -
-
-
- -
-
-
- -
-
-
91.5 -
-
-
90 75.7
75.7
83.7
85 -
-
-
- -
-
-
- -
-
-
- 36.7
36.7
55
50 51.1
51.1
65.6
- 23.6
23.6
37.2
- 22.4
22.4
36.2
- -
-
-
30 -
-
-
37 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | AIP - O00170 |
| 2 | MYBPC3 - Q14896 |
| 3 | ACTC1 - P68032 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
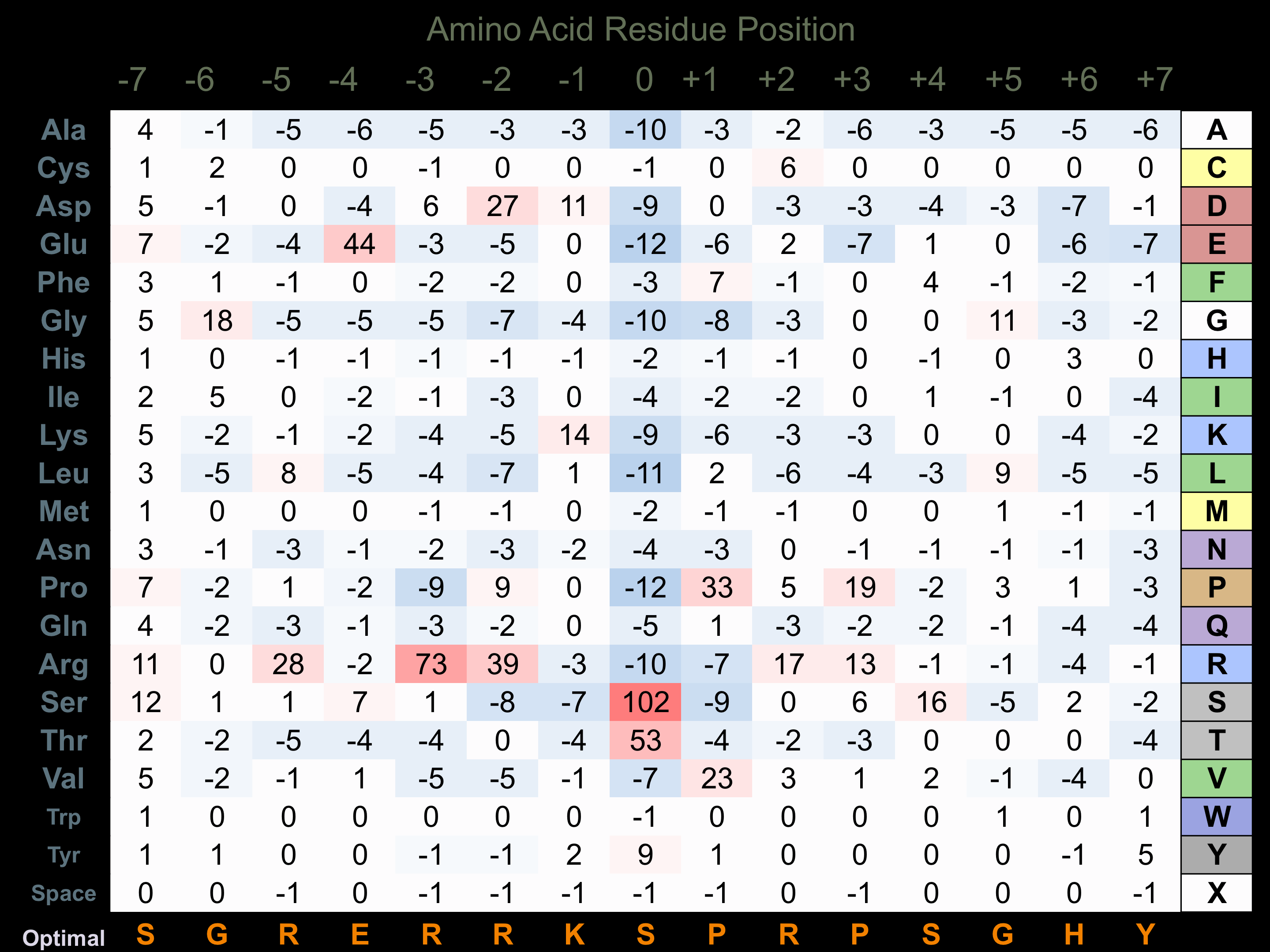
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cardiovascular disorders
Specific Diseases (Non-cancerous):
Restrictive cardiomyopathy
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for HH498 in diverse human cancers of 297, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 15 for this protein kinase in human cancers was 0.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 24979 diverse cancer specimens. This rate is 1.62-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.67 % in 864 skin cancers tested; 0.49 % in 1297 large intestine cancers tested; 0.4 % in 589 stomach cancers tested; 0.26 % in 1823 lung cancers tested; 0.25 % in 603 endometrium cancers tested; 0.24 % in 710 oesophagus cancers tested; 0.22 % in 238 bone cancers tested; 0.12 % in 548 urinary tract cancers tested; 0.08 % in 881 prostate cancers tested; 0.07 % in 1512 liver cancers tested; 0.05 % in 1459 pancreas cancers tested; 0.05 % in 1276 kidney cancers tested; 0.04 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E662K (5).
Comments:
Ten deletions, no insertions or complex mutations are noted on the COSMIC website.

