Nomenclature
Short Name:
HIPK2
Full Name:
Homeodomain-interacting protein kinase 2
Alias:
- EC 2.7.11.1
- Sialophorin tail associated nuclear serine,threonine kinase
- Sialophorin tail associated nuclear serine/threonine kinase
- STANK
- Homeodomain interacting protein kinase 2
- Kinase HIPK2
- NBAK1
- Nuclear body associated kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
DYRK
SubFamily:
HIPK
Specific Links
Structure
Mol. Mass (Da):
130,966
# Amino Acids:
1198
# mRNA Isoforms:
3
mRNA Isoforms:
130,966 Da (1198 AA; Q9H2X6); 128,158 Da (1171 AA; Q9H2X6-3); 101,013 Da (918 AA; Q9H2X6-2)
4D Structure:
Interacts with TRADD, TP53, TP73, TP63, CREBBP, DAXX, P53DINP1, SKI, SMAD1, SMAD2 and SMAD3, but not SMAD4. Interacts with NKX1-2, NKX2-5, SPN/CD43, UBE2I, HMGA1, CTBP1, AXIN1, NLK, MYB, POU4F1, POU4F2, POU4F3, UBE2I, UBL1. Probably part of a complex consisting of TP53, HIPK2 and AXIN1 By similarity. Interacts with CBX4
1D Structure:
Subfamily Alignment
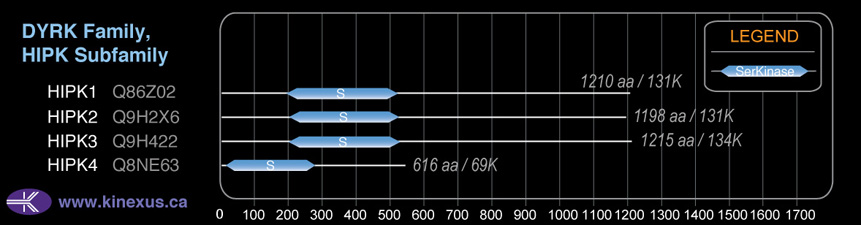
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 199 | 527 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S46, S121, S336, S359, S364-, S668, S826, S827, S848, S882, S924, S934.
Sumoylated:
K32, K1191.
Threonine phosphorylated:
T119, T221, T360, T796, T838, T880, T1114.
Tyrosine phosphorylated:
Y44, Y258, Y264, Y340, Y361, Y366, Y367, Y423, Y443, Y558, Y1136, Y1197.
Ubiquitinated:
K32, K430, K552, K565, K671, K803, K805, K1191.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1450
80
1029
 10
10
142
21
202
 44
44
637
14
1280
 48
48
692
309
1093
 60
60
867
103
653
 5
5
67
154
108
 8
8
113
111
208
 71
71
1025
65
2158
 30
30
442
17
340
 39
39
559
288
842
 47
47
687
45
3423
 57
57
820
236
751
 14
14
207
36
487
 3
3
41
9
37
 31
31
451
39
1397
 11
11
153
54
188
 46
46
668
430
3309
 30
30
428
21
656
 18
18
261
233
426
 45
45
647
333
575
 60
60
869
37
1865
 17
17
243
41
508
 78
78
1124
16
2013
 21
21
305
21
866
 14
14
204
37
348
 71
71
1031
197
1509
 15
15
218
39
528
 59
59
856
21
2491
 28
28
407
21
1144
 15
15
221
140
317
 58
58
845
24
800
 27
27
390
88
772
 15
15
213
197
661
 74
74
1069
260
809
 14
14
201
139
350
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.3
100 97
97
97.3
99.5 -
-
-
- -
-
-
- 65.4
65.4
76.4
98 -
-
-
- 96.7
96.7
98
97 60.9
60.9
71.9
97 -
-
-
- -
-
-
- 92.9
92.9
95.4
94 23.5
23.5
35.7
83 80
80
85.8
82 -
-
-
- 22
22
37.3
46 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
38 -
-
-
40
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TP53 - P04637 |
| 2 | UBE2I - P63279 |
| 3 | SUMO1 - P63165 |
| 4 | HMGA1 - P17096 |
| 5 | TP53INP1 - Q96A56 |
| 6 | WSB1 - Q9Y6I7 |
| 7 | MYB - P10242 |
| 8 | SP100 - P23497 |
| 9 | NKX3-1 - Q99801 |
| 10 | DAXX - Q9UER7 |
| 11 | NKX2-1 - P43699 |
| 12 | PAX6 - P26367 |
| 13 | TRADD - Q15628 |
| 14 | CREBBP - Q92793 |
| 15 | RANBP9 - Q96S59 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
By UV.
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AML1 (RUNX1) | Q01196 | S249 | DTRQIQPSPPWSYDQ | + |
| AML1 (RUNX1) | Q01196 | S276 | VHPATPISPGRASGM | |
| AML1 (RUNX1) | Q01196 | T273 | SPSVHPATPISPGRA | + |
| CBX4 | O00257 | T495 | SLQVKPETPASAAVA | + |
| CTBP1 | Q13363 | S422 | AHPPHAPSPGQTVKP | ? |
| HIPK2 | Q9H2X6 | Y361 | SKAVCSTYLQSRYYR | + |
| p27Kip1 | P46527 | S10 | NVRVSNGSPSLERMD | - |
| p53 | P04637 | S46 | AMDDLMLSPDDIEQW | + |
| PAX6 | P26367 | T281 | QRRQASNTPSHIPIS | |
| PAX6 | P26367 | T304 | QPIPQPTTPVSSFTS | + |
| PAX6 | P26367 | T373 | GRSYDTYTPPHMQTH | |
| PML | P29590 | S36 | PSEGRQPSPSPSPTE | + |
| PML | P29590 | S38 | EGRQPSPSPSPTERA | + |
| PML | P29590 | S8 | MEPAPARSPRPQQDP | |
| SIAH2 | O43255 | S28 | PQPQHTPSPAAPPAA | |
| STAT3 | P40763 | S727 | NTIDLPMSPRTLDSL | - |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
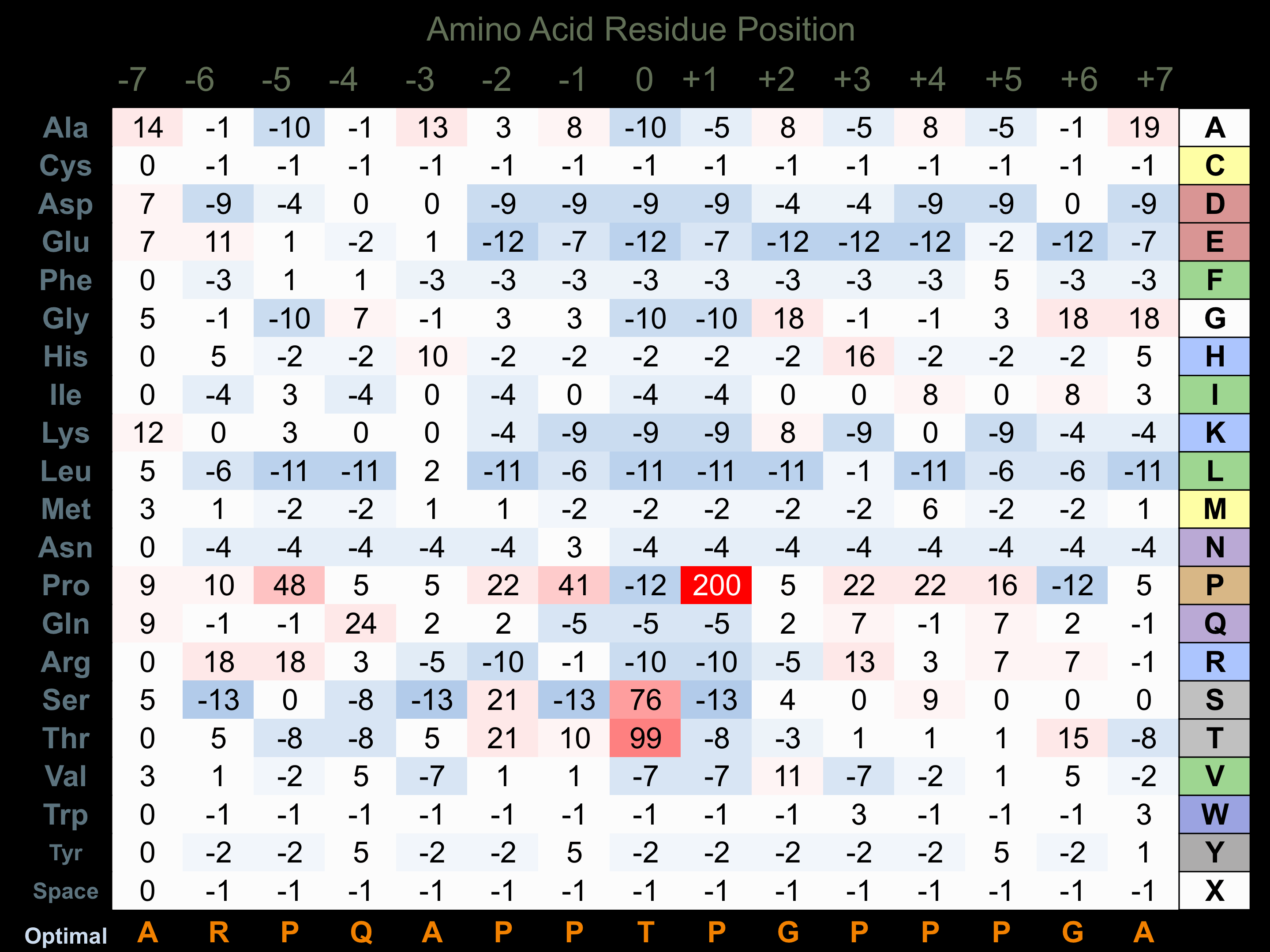
Matrix Type:
Experimentally derived from alignment of 33 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Familial adenomatous polyposis (FAP)
Comments:
HIPK2 may be an oncoprotein (OP) or a tumour suppressor protein (TSP). HIPK2 is upregulated in some cervical cancers, and down regulated in some thyroid and breast carcinomas. Functional studies must be performed before committing HIPK2 to the tumour suppressor proteins or oncoproteins class of tumour mediators. Location to the nucleoplasm can be erroneously induced and kinase activity inhibited towards PML, RUNX1, and EP300 (but not RANBP9) with a K228A mutation. Enzymatic activity can be inhibited, without affecting BMP-induced transcriptional activation, or complexing with TP53/TP73 with a K228R mutation. BMP-induced transcriptional activation is increased when the K228R mutation is in conjunction with S359A + T360A + Y361F. Nuclear localization is inhibited with the K803A + K805A, or R833A, or K835E mutations. The V885K + S886F + V887M + I 888H + T889F + I890H + S891R + S892M mutations or the D893N + T894F + D895N+ E896Q + E897Q + E898Q + E899Q mutations lead to inhibited SUMO and CBX4 interaction, alongside reduced nuclear and PML-nuclear bodies localization of HIPK2. Loss of SUMO interaction and impaired PML-nuclear body localization occured with the V885K + S886S + V887A + I888K or with the S892A + D893D + T894T + D895A mutations.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +480, p<0.0002); Ovary adenocarcinomas (%CFC= +46, p<0.055); Prostate cancer - primary (%CFC= +47, p<0.001); Skin fibrosarcomas (%CFC= +105, p<0.052); Skin melanomas - malignant (%CFC= +50, p<0.001); Skin squamous cell carcinomas (%CFC= +68, p<0.003); and Uterine fibroids (%CFC= +56, p<0.0004). The COSMIC website notes an up-regulated expression score for HIPK2 in diverse human cancers of 465, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 9 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25486 diverse cancer specimens. This rate is only -10 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 1093 large intestine cancers tested; 0.27 % in 805 skin cancers tested; 0.21 % in 602 endometrium cancers tested; 0.18 % in 589 stomach cancers tested; 0.09 % in 1619 lung cancers tested; 0.09 % in 1292 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: V85A (5).
Comments:
Only 5 deletions, 2 insertions and no complex mutations are noted on the COSMIC website.

