Nomenclature
Short Name:
IKKe
Full Name:
Inhibitor of nuclear factor kappa-B kinase epsilon subunit
Alias:
- EC 2.7.11.10
- KIAA0151
- I kappa-B kinase epsilon
- IkBKE
- IKKE
- IKKI
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
IKK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
80,462
# Amino Acids:
716
# mRNA Isoforms:
2
mRNA Isoforms:
80,462 Da (716 AA; Q14164); 70,816 Da (631 AA; Q14164-2)
4D Structure:
May interact with MAVS/IPS1. Interacts with AZI2. Interacts with SIKE1. Interacts with TICAM1/TRIF, IRF3 and DDX58/RIG-I, interactions are disrupted by the interaction between IKBKE and SIKE1. Interacts with TOPORS; induced by DNA damage. Interacts with CYLD.
1D Structure:
Subfamily Alignment
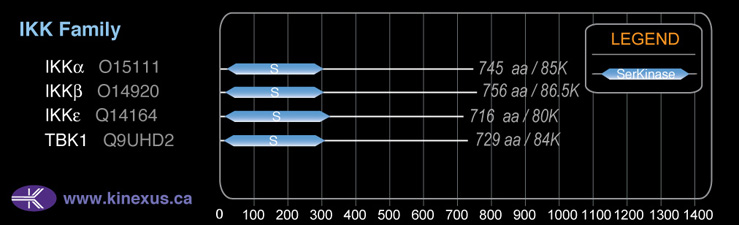
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 9 | 311 | Pkinase |
| 436 | 457 | Leucine zipper |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S172+, S464, S479, S522, S664.
Sumoylated:
K231.
Threonine phosphorylated:
T463, T474, T501.
Tyrosine phosphorylated:
Y179, Y671.
Ubiquitinated:
K30, K137, K401.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 48
48
988
29
1208
 3
3
58
15
45
 5
5
112
23
117
 13
13
261
98
457
 25
25
514
25
428
 2
2
42
74
21
 11
11
226
31
492
 20
20
421
48
600
 11
11
224
17
159
 5
5
98
94
123
 6
6
131
42
186
 30
30
609
184
617
 7
7
146
45
131
 3
3
66
12
43
 6
6
119
36
123
 2
2
44
15
14
 4
4
85
130
64
 7
7
140
32
139
 4
4
78
105
56
 20
20
404
109
416
 4
4
79
33
70
 6
6
132
38
97
 9
9
195
33
151
 3
3
69
32
75
 10
10
206
34
177
 22
22
447
64
560
 6
6
125
48
95
 6
6
123
31
105
 4
4
90
32
94
 23
23
467
28
175
 41
41
846
24
755
 100
100
2063
36
4245
 29
29
595
73
1076
 40
40
834
57
679
 3
3
72
44
68
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 43.9
43.9
55
97 97.3
97.3
98.9
- -
-
-
88 -
-
-
85 79.9
79.9
83.6
90.5 -
-
-
- 82.6
82.6
91.1
84 20.7
20.7
39.6
84 -
-
-
- 50.6
50.6
58.9
- 21.7
21.7
40.7
64 47.9
47.9
64.3
58 21.5
21.5
39.7
50 -
-
-
- 23.2
23.2
38.5
- 37.4
37.4
56.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | TANK - Q92844 |
| 2 | NFKBIA - P25963 |
| 3 | CYLD - Q9NQC7 |
| 4 | TICAM1 - Q8IUC6 |
| 5 | IRF3 - Q14653 |
| 6 | CLTC - Q00610 |
| 7 | MYH10 - P35580 |
| 8 | MYL12A - P19105 |
| 9 | CDC37 - Q16543 |
| 10 | CALM1 - P62158 |
| 11 | RUVBL1 - Q9Y265 |
| 12 | TPM3 - P06753 |
| 13 | UBE2N - P61088 |
| 14 | DDX5 - P17844 |
Regulation
Activation:
Phosphorylation at Ser-172 increases phosphotransferase activity.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| IkBa | P25963 | S32 | LLDDRHDSGLDSMKD | - |
| IkBa | P25963 | S36 | RHDSGLDSMKDEEYE | - |
| IKKe (IKBKE) | Q14164 | S172 | DDDEKFVSVYGTEEY | + |
| IRF3 | Q14653 | S386 | ARVGGASSLENTVDL | + |
| IRF3 | Q14653 | S396 | NTVDLHISNSHPLSL | + |
| IRF3 | Q14653 | S398 | VDLHISNSHPLSLTS | + |
| IRF3 | Q14653 | S402 | ISNSHPLSLTSDQYK | ? |
| IRF7 | Q92985 | S471 | GTQREGVSSLDSSSL | |
| IRF7 | Q92985 | S472 | TQREGVSSLDSSSLS | - |
| IRF7 | Q92985 | S477 | VSSLDSSSLSLCLSS | + |
| IRF7 | Q92985 | S479 | SLDSSSLSLCLSSAN | + |
| NFkB-p65 | Q04206 | S468 | AVFTDLASVDNSEFQ | |
| NFkB-p65 | Q04206 | S536 | SGDEDFSSIADMDFS | |
| STAT1 | P42224 | S708 | YIKTELISVSEVHPS | + |
| STAT1 | P42224 | S745 | VGSVEFDSMMNTV |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
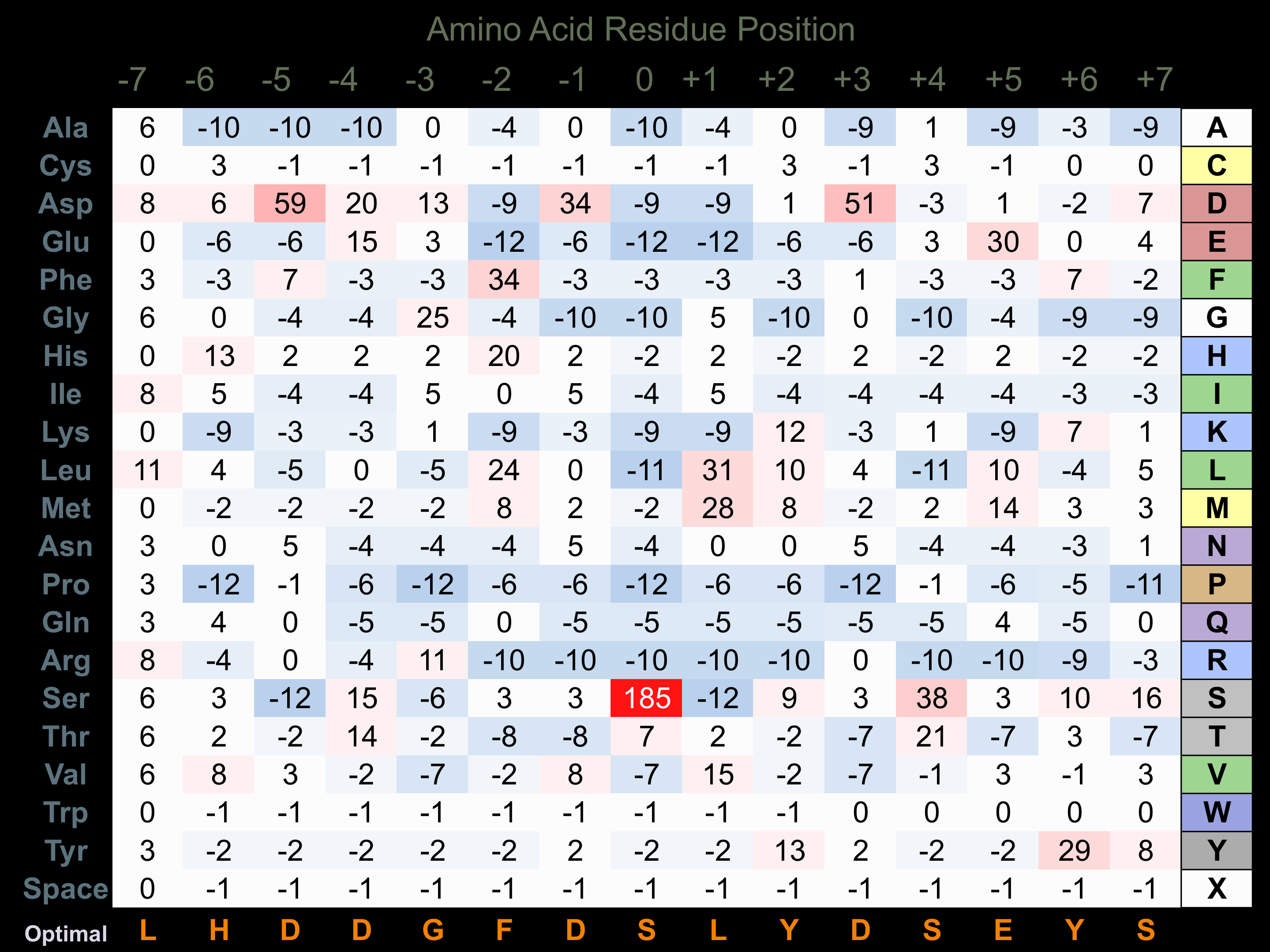
Matrix Type:
Experimentally derived from alignment of 28 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, autoimmune disorders
Specific Diseases (Non-cancerous):
Sle susceptibility
Comments:
Sle Susceptibility is related to lupus erythematosus and systemic lupus erythematosus where there is an autoimmune response that can attack joints, skin, blood vessels, or organs resulting in systemic inflammation. Ubiquitination and decreased kinase phosphotransferase activity stems from a K30A, or K401A, or K30R, or K401R mutation in IKKe. K30R and K401R will additionally reduce interactions with IKKb, IKBKG, and MyD88. The K38A mutation leads to kinase phosphotransferase activity inhibition and inhibited nuclear transport. The E168A, or S172E mutations will partially decrease kinase phosphotransferase activity. A S172A mutation prevents autophosphorylation and leads to impaired kinase phosphotransferase activity. Sumoylation and targeting to nuclear bodies is lost with a K231R mutation to IKKe.
Specific Cancer Types:
Breast cancer
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -50, p<0.081); Classical Hodgkin lymphomas (%CFC= +50, p<0.0007); Large B-cell lymphomas (%CFC= +85, p<0.008); Lung adenocarcinomas (%CFC= +65, p<0.0001); Ovary adenocarcinomas (%CFC= +143, p<0.0002); and Uterine leiomyosarcomas (%CFC= +70, p<0.018). The COSMIC website notes an up-regulated expression score for IKBKE in diverse human cancers of 492, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 22 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. Ubiquitination and decreased kinase phosphotransferase activity can arise from a K30A, K401A, K30R, or K401R mutation in IKKe. K30R and K401R will additionally reduce interactions with IKKb, IKBKG, and MyD88. The K38A mutation can lead to kinase phosphotransferase activity inhibition and inhibited nuclear transport. The E168A or S172E mutations can partially decrease kinase phosphotransferase activity. A S172A mutation can prevent autophosphorylation and lead to impaired kinase phosphotransferase activity. Sumoylation and targeting to nuclear bodies can be lost with a K231R mutation to IKKe.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25518 diverse cancer specimens. This rate is very similar (+ 7% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.47 % in 805 skin cancers tested; 0.38 % in 1093 large intestine cancers tested; 0.28 % in 589 stomach cancers tested; 0.23 % in 602 endometrium cancers tested; 0.1 % in 1942 lung cancers tested; 0.09 % in 1488 breast cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,736 cancer specimens
Comments:
Only 4 deletions, 2 insertions, and no complex mutations are noted on the COSMIC website.

