Nomenclature
Short Name:
IRE2
Full Name:
Serine-threonine-protein kinase-endoribonuclease IRE2
Alias:
- Inositol-requiring 2
- ERN2
- hIRE2p
- IRE1b
- Ire1beta
- Endoplasmic reticulum-to-nucleus signaling 2
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
IRE
SubFamily:
NA
Structure
Mol. Mass (Da):
102466
# Amino Acids:
926
# mRNA Isoforms:
1
mRNA Isoforms:
102,480 Da (926 AA; Q76MJ5)
4D Structure:
NA
1D Structure:
Subfamily Alignment
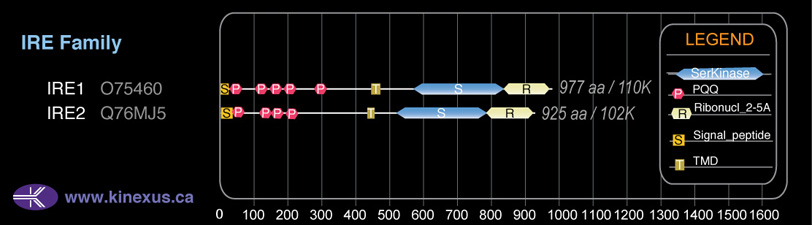
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 34 | signal_peptide |
| 35 | 65 | PQQ |
| 115 | 147 | PQQ |
| 148 | 180 | PQQ |
| 192 | 223 | PQQ |
| 431 | 451 | TMD |
| 520 | 781 | Pkinase |
| 785 | 911 | Ribonuc_2-5A |
| 784 | 912 | KEN |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S268, S802.
Tyrosine phosphorylated:
Y860.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 75
75
1138
22
1075
 4
4
57
6
38
 22
22
327
21
890
 40
40
607
83
1697
 49
49
737
21
508
 3
3
45
46
35
 16
16
245
22
235
 91
91
1385
34
2582
 73
73
1111
7
718
 2
2
36
52
27
 4
4
55
23
89
 54
54
816
85
696
 4
4
67
33
113
 2
2
32
3
12
 9
9
140
24
227
 4
4
59
12
24
 6
6
92
35
139
 14
14
212
24
659
 4
4
58
73
79
 37
37
565
81
496
 19
19
287
24
974
 7
7
99
25
232
 12
12
183
23
157
 6
6
91
24
154
 11
11
170
24
562
 94
94
1424
60
3872
 6
6
94
33
241
 22
22
327
24
408
 17
17
251
24
897
 11
11
174
28
167
 25
25
376
18
254
 100
100
1519
26
1985
 1
1
22
29
16
 56
56
844
52
708
 5
5
72
26
59
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 94.2
94.2
94.4
99 77.6
77.6
79.5
96 -
-
-
86 -
-
-
- 84.2
84.2
88.9
85 -
-
-
- 80.5
80.5
86.2
83 -
-
-
82 -
-
-
- 53.5
53.5
61.8
- -
-
-
64 -
-
-
58 -
-
-
- -
-
-
- -
-
-
- 38.4
38.4
54.3
- 37.4
37.4
54.5
- 36
36
48.2
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 27.6
27.6
44.2
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SEC13 - P55735 |
| 2 | COPB2 - P35606 |
Regulation
Activation:
The kinase domain is activated by trans-autophosphorylation. Kinase activity is required for activation of the endoribonuclease domain
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
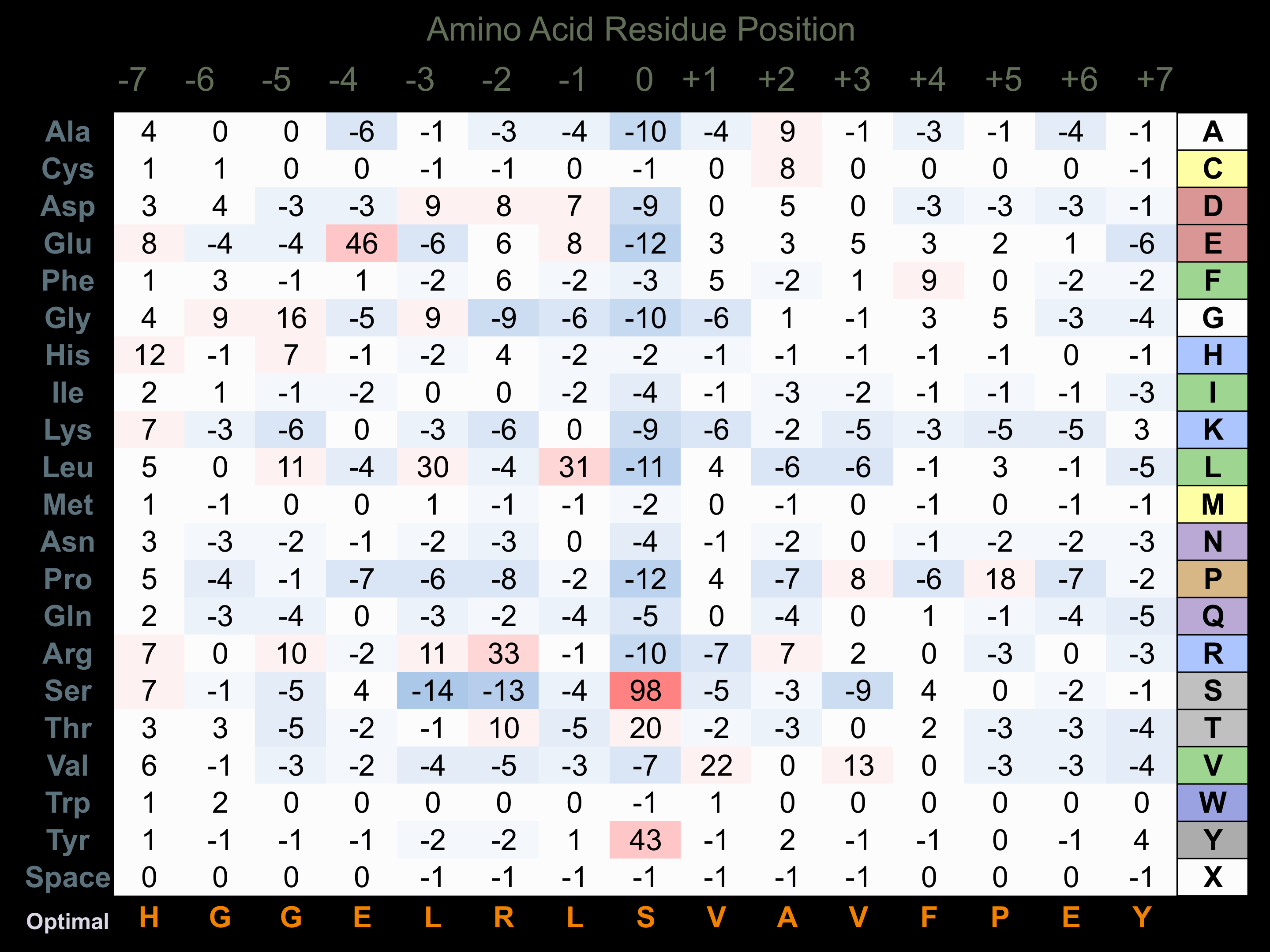
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Gastrointestinal disorders
Specific Diseases (Non-cancerous):
Inflammatory bowel disease (IBD)
Comments:
Inflammatory bowel disease (IBD) is a gastrointestinal disease characterized by recurrent inflammation of the bowels, leading to gastrointestinal distress. The general term IBD encompasses the inflammatory disorders Crohn's disease and ulcerative colitis. IBD is thought to be caused by over-exaggerated inflammatory responses to commensal microbes found in the gastrointestinal tract. The occurence of IBD shows a strong familial component, indicating that genetic factors may be important contributors to the disease pathogenesis. In animal models, mice lacking IRE2 expression display increased ER stress in intestinal epithelial cells and significantly increased occurence of colitis compared to wild-type controls. This suggests that IRE2 loss-of-function and increased ER stress in the intestinal epithelium may contribute to the pathogenesis of IBD.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.1 % in 24449 diverse cancer specimens. This rate is only 36 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.54 % in 864 skin cancers tested; 0.32 % in 943 upper aerodigestive tract cancers tested; 0.31 % in 1229 large intestine cancers tested; 0.23 % in 603 endometrium cancers tested; 0.23 % in 1609 lung cancers tested; 0.18 % in 548 urinary tract cancers tested; 0.17 % in 569 stomach cancers tested; 0.12 % in 710 oesophagus cancers tested; 0.08 % in 1512 liver cancers tested; 0.08 % in 1289 breast cancers tested; 0.06 % in 881 prostate cancers tested; 0.06 % in 1437 pancreas cancers tested; 0.05 % in 441 autonomic ganglia cancers tested; 0.03 % in 807 ovary cancers tested; 0.03 % in 1982 haematopoietic and lymphoid cancers tested; 0.03 % in 1253 kidney cancers tested; 0.02 % in 2030 central nervous system cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: T202P (21).
Comments:
Only 8 deletions, and no insertions mutations or complex mutations are noted on the COSMIC website.

