Nomenclature
Short Name:
JAK3
Full Name:
Tyrosine-protein kinase JAK3
Alias:
- EC 2.7.10.2
- Kinase Jak3
- Leukocyte janus kinase
- LJAK
- L-JAK
- JAK-3
- JAK3_HUMAN
- JAKL
- Janus kinase 3
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
JakA
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
125,099
# Amino Acids:
1124
# mRNA Isoforms:
3
mRNA Isoforms:
125,099 Da (1124 AA; P52333); 121,268 Da (1094 AA; P52333-2); 68,279 Da (619 AA; P52333-4)
4D Structure:
Interacts with STAM2 and MYO18A By similarity. Interacts with SHB.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
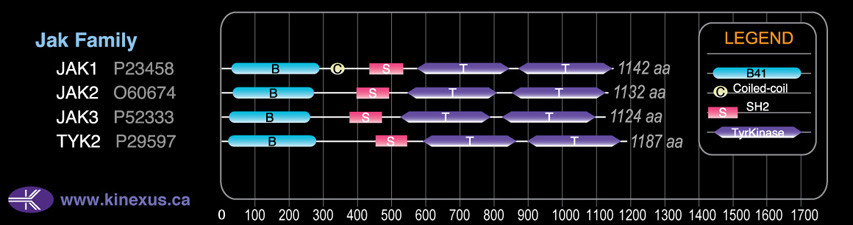
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K748.
Serine phosphorylated:
S5, S15, S17, S20, S398, S498, S568, S663, S782, S931, S1120.
Threonine phosphorylated:
T8+, T21, T391+.
Tyrosine phosphorylated:
Y506, Y536, Y785+, Y904+, Y929, Y939+, Y980+, Y981+.
Ubiquitinated:
K122, K157, K361, K734, K823, K972.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 43
43
633
48
949
 4
4
67
25
111
 5
5
69
26
116
 13
13
191
171
341
 33
33
494
44
398
 1.3
1.3
20
120
26
 26
26
381
65
629
 33
33
490
72
930
 21
21
319
24
230
 9
9
130
186
152
 6
6
85
63
117
 30
30
442
318
530
 8
8
126
59
163
 4
4
58
18
81
 4
4
67
50
102
 3
3
41
29
50
 5
5
71
460
111
 8
8
120
38
157
 4
4
67
170
85
 20
20
304
190
312
 5
5
71
50
110
 10
10
146
57
326
 7
7
105
37
119
 18
18
267
39
1304
 10
10
150
49
261
 38
38
570
107
1017
 6
6
89
62
113
 11
11
170
40
640
 4
4
57
37
92
 12
12
180
56
225
 26
26
394
30
444
 100
100
1489
62
4085
 10
10
149
102
386
 46
46
691
109
645
 6
6
89
61
118
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 20.6
20.6
35.9
100 83.1
83.1
83.4
97 -
-
-
90 -
-
-
96 88.6
88.6
92.1
90 -
-
-
- 82.3
82.3
87.9
84 80.1
80.1
86.4
83 -
-
-
- 42.7
42.7
59.1
- 50.1
50.1
68.1
61.5 48.8
48.8
66.9
54 39
39
57.3
52 -
-
-
- 22.6
22.6
40.8
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Phosphorylation at Thr-8, Tyr-904 and Tyr-980 increases phosphotransferase activity. Phosphorylation at Tyr-939 increases phosphotransferase activity and induces interaction with STAT5A. Phosphorylation at Tyr-785 induces interaction with SH2-B-beta.
Inhibition:
Phosphorylation at Tyr-981 inhibits phosphotransferase activity.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| JAK1 | P23458 | Y1022 | AIETDKEYYTVKDDR | |
| JAK1 | P23458 | Y1023 | IETDKEYYTVKDDRD | |
| JAK1 | P23458 | Y208 | KDISYKRYIPETLNK | |
| JAK3 | P52333 | T8 | MAPPSEETPLIPQRS | + |
| JAK3 | P52333 | Y785 | NSLISSDYELLSDPT | + |
| JAK3 | P52333 | Y904 | SLRLVMEYLPSGCLR | + |
| JAK3 | P52333 | Y939 | QICKGMEYLGSRRCV | + |
| JAK3 | P52333 | Y980 | LLPLDKDYYVVREPG | + |
| JAK3 | P52333 | Y981 | LPLDKDYYVVREPGQ | + |
| RACK1 | P63244 | Y194 | NHIGHTGYLNTVTVS | |
| SH2-Bb | Q9NRF2 | Y439 | DRLSQGAYGGLSDRP | |
| SH2-Bb | Q9NRF2 | Y494 | VHPLSAPYPPLDTPE | |
| SIGLEC10 | Q96LC7 | Y597 | RHSTILDYINVVPTA | |
| SIGLEC10 | Q96LC7 | Y667 | ESQEELHYATLNFPG | |
| SIGLEC10 | Q96LC7 | Y691 | PKGTQADYAEVKFQ_ | |
| STAT1 | P42224 | Y701 | DGPKGTGYIKTELIS | + |
| STAT5A | P42229 | Y694 | LAKAVDGYVKPQIKQ | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
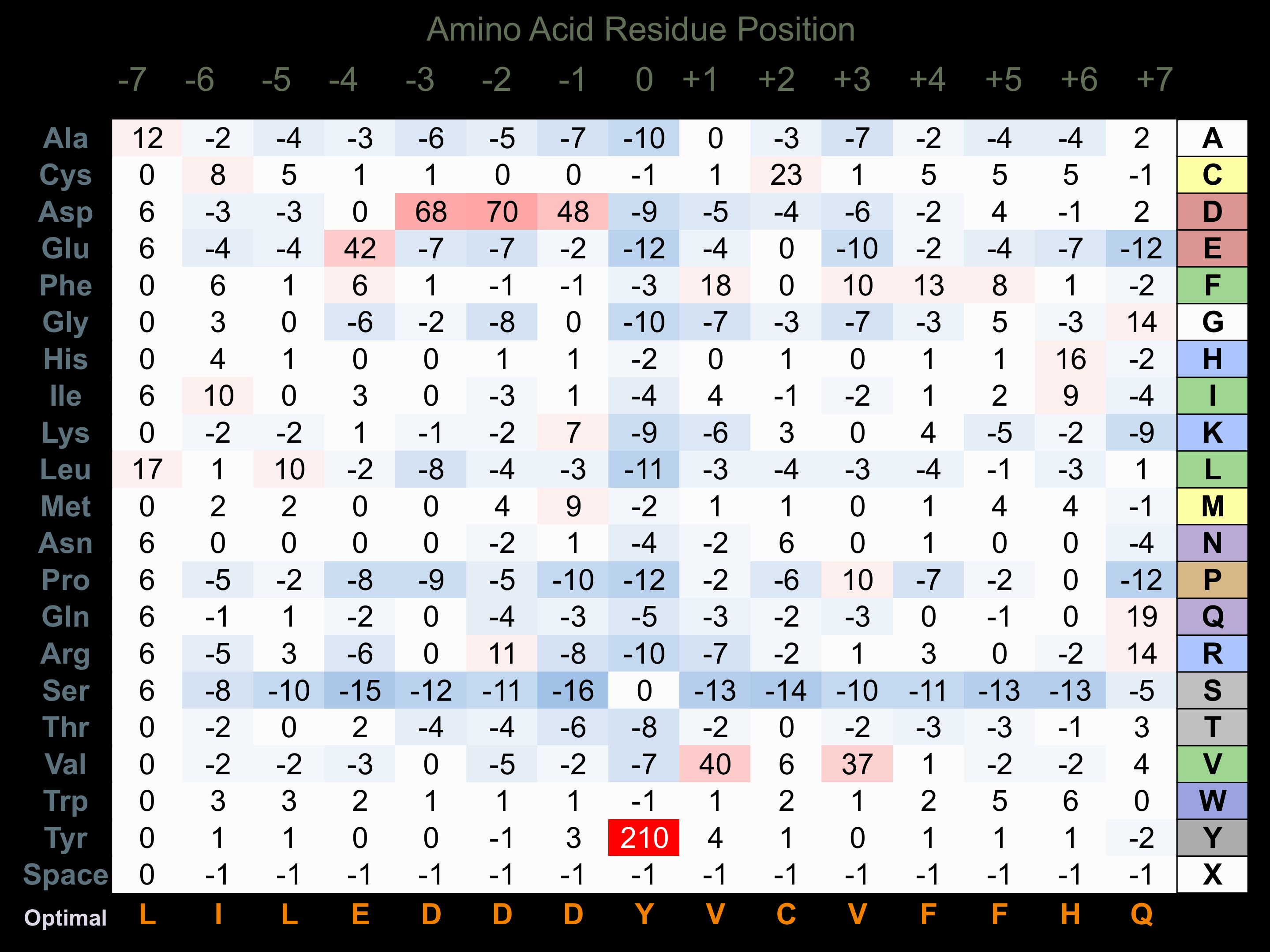
Matrix Type:
Experimentally derived from alignment of 13 known protein substrate phosphosites and 48 peptides phosphorylated by recombinant JAK3 in vitro tested in-house by Kinexus.
Domain #:
2
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, immune disorders
Specific Diseases (Non-cancerous):
Scid, autosomal recessive, T-negative/b-Positive Type; Severe combined immunodeficiency; Jacobsen syndrome; Severe combined Immune deficiency, autosomal recessive, T cell-negative, B Cell-Positive, NK Cell-negative, JAK3-related
Comments:
Mutations at many sites in JAK3 are associated with severe combined immunodeficiency autosomal recessive T-cell-negative/B-cell-positive/NK-cell-negative. It is a form or severe combined immunodeficiency (SCID), and characterized by both humoral and cell-mediated immunity impairment, leukopenia, and low or absent antibody levels, leading to infancy recurrent, persistent infections by opportunistic organisms.
Specific Cancer Types:
Anaplastic large cell lymphomas (ALCL)
Comments:
JAK3 may be an oncoprotein (OP). JAK3 mutations were commonly found in juvenile myelomonocytic leukemia from whole-exome sequencing studies from 13 individuals. JAK3 were hypothesized to be involved in the progression rather than the initiation of leukemia, and patients with mutated JAK3 were associated with poorer clinical results.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= -51, p<0.028); Clear cell renal cell carcinomas (cRCC) (%CFC= +136, p<0.007); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +675, p<0.006); Gastric cancer (%CFC= -60, p<0.005); Large B-cell lymphomas (%CFC= +87, p<0.033); andProstate cancer - primary (%CFC= +181, p<0.0001). The COSMIC website notes an up-regulated expression score for JAK3 in diverse human cancers of 331, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 30293 diverse cancer specimens. This rate is a modest 1.56-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.42 % in 1240 large intestine cancers tested; 0.32 % in 809 skin cancers tested; 0.25 % in 4940 haematopoietic and lymphoid cancers tested; 0.17 % in 903 stomach cancers tested; 0.16 % in 603 endometrium cancers tested; 0.12 % in 984 upper aerodigestive tract cancers tested; 0.11 % in 2093 lung cancers tested; 0.1 % in 1582 breast cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: A572V (27); A657Q (26); M511T (22); A573V (16).
Comments:
Only 7 deletions, 4 insertions and no complex mutations are noted on the COSMIC website.

