Nomenclature
Short Name:
KIS
Full Name:
Serine/threonine-protein kinase Kist
Alias:
- UHMK1
- BC014917
- KIST
- U2AF homology motif kinase 1
- Kinase interacting with stathmin
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
Other-Unique
SubFamily:
NA
Structure
Mol. Mass (Da):
46,546
# Amino Acids:
419
# mRNA Isoforms:
3
mRNA Isoforms:
46,546 Da (419 AA; Q8TAS1); 39,044 Da (345 AA; Q8TAS1-3); 38,130 Da (344 AA; Q8TAS1-2)
4D Structure:
Interacts with stathmin and CDKN1B/p27Kip1
1D Structure:
Subfamily Alignment
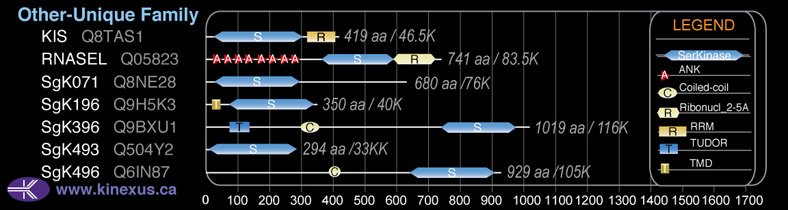
Domain Distribution:
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S283, S290.
Tyrosine phosphorylated:
Y197.
Ubiquitinated:
K190, K282, K383.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 35
35
1572
24
1084
 21
21
921
8
1462
 -
-
-
-
-
 7
7
309
84
727
 19
19
852
29
565
 2
2
81
27
60
 1
1
58
33
44
 100
100
4429
15
6963
 0.3
0.3
15
3
0
 6
6
282
59
440
 12
12
519
11
897
 19
19
862
40
659
 14
14
614
2
7
 13
13
597
5
894
 6
6
268
11
367
 5
5
222
17
482
 7
7
321
85
439
 9
9
391
5
552
 8
8
354
41
1203
 22
22
987
84
749
 7
7
308
11
431
 5
5
208
9
384
 -
-
-
-
-
 28
28
1254
7
1179
 20
20
890
11
1336
 29
29
1306
56
2662
 12
12
542
5
713
 23
23
1034
5
1386
 18
18
801
5
1122
 6
6
259
28
108
 25
25
1101
12
53
 27
27
1190
23
2685
 7
7
289
71
550
 21
21
921
78
642
 3
3
120
57
80
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.7
99.7
100
100 99.7
99.7
100
100 -
-
-
100 -
-
-
- 78.2
78.2
78.2
100 -
-
-
- 99.2
99.2
99.5
99 99.2
99.2
99.7
99 -
-
-
- -
-
-
- -
-
-
88 -
-
-
- 71.5
71.5
83
74 -
-
-
- 21.2
21.2
36
- -
-
-
- 20.5
20.5
37.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PAM - P19021 |
| 2 | STMN1 - P16949 |
| 3 | ABL1 - P00519 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
By serum growth factors
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
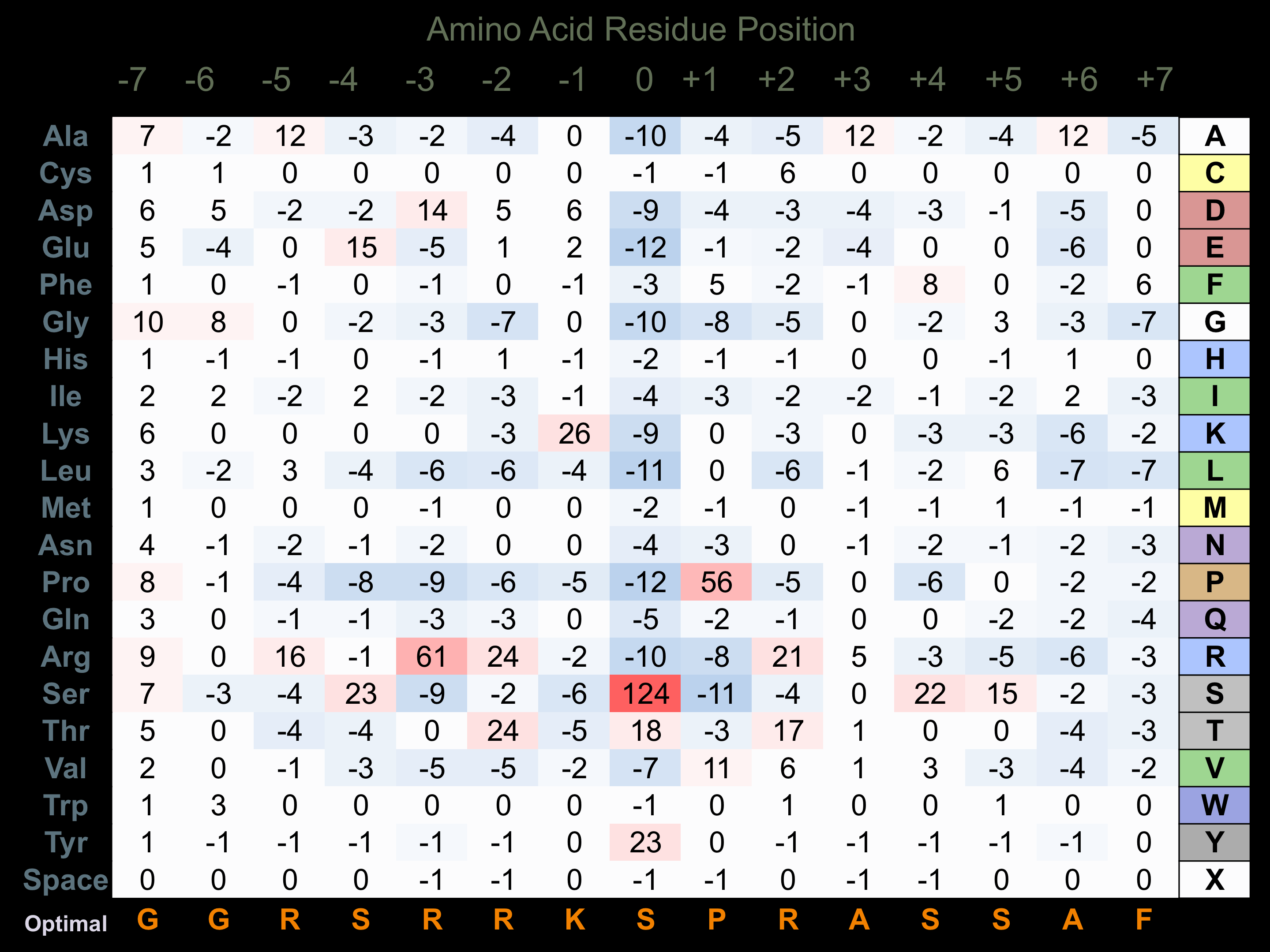
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Neurological disorders
Specific Diseases (Non-cancerous):
Schizophrenia
Comments:
The KIS gene is located in the 1q23.3 chromosome region and has been implicated as a susceptibility gene for schizophrenia, as different gene variants have been linked with the occurence of the disease. For example, three SNPs from schizophrenia patients in Aberdeen, Scotland and London, England (University College London) (rs7513662, rs6427680, rs6694863), as well as several haplotypes (e.g. HAP-A, HAP-B, HAP-C, etc.) were identified to be signficantly associated with schizophrenia. As none of these SNPs are located within the coding region of the KIS gene, they are predicted to potentially confer susceptibility by affecting KIS expression levels. However, post-mortem analysis of tissue samples from the superior temporal gyrus, dorsolateral prefrontal cortex, anterior cingulate cortex, and cerebellum of schizophrenia patients did not reveal any difference in KIS expression (either protein or mRNA) as compared to control patients.
Comments:
Significantly elevated KIS expression is observed in NF1-plexiform neurofibromas and malignant peripheral nerve sheath tumours.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for KIS in diverse human cancers of 662, which is 1.4-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 9 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24751 diverse cancer specimens. This rate is only -34 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.28 % in 589 stomach cancers tested; 0.24 % in 603 endometrium cancers tested; 0.17 % in 273 cervix cancers tested; 0.15 % in 1634 lung cancers tested; 0.15 % in 1270 large intestine cancers tested; 0.14 % in 864 skin cancers tested; 0.12 % in 382 soft tissue cancers tested; 0.09 % in 548 urinary tract cancers tested; 0.05 % in 881 prostate cancers tested; 0.05 % in 1512 liver cancers tested; 0.03 % in 710 oesophagus cancers tested; 0.02 % in 1316 breast cancers tested; 0.02 % in 1276 kidney cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
None > 2 in 20,034 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

