Nomenclature
Short Name:
MEKK4
Full Name:
Mitogen-activated protein kinase kinase kinase 4
Alias:
- EC 2.7.11.25
- KIAA0213
- MAPK/ERK kinase kinase 4
- MAPKKK4
- MEK kinase 4
- MTK1
- Kinase MEKK4
- M3K4
- MAP three kinase 1
- MAP3K4
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE11
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
181685
# Amino Acids:
1608
# mRNA Isoforms:
2
mRNA Isoforms:
181,685 Da (1608 AA; Q9Y6R4); 177,014 Da (1558 AA; Q9Y6R4-2)
4D Structure:
Binds both upstream activators and downstream substrates in multimolecular complexes. Interacts with AXIN1 and DIXDC1; interaction with DIXDC1 prevents interaction with AXIN1.
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1343 | 1601 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K98, K890.
Serine phosphorylated:
S63, S66, S72, S84, S87, S173, S253, S431, S435, S456, S457, S461, S481, S499, S798, S835, S979, S1068, S1135, S1252, S1268, S1270, S1274, S1282.
Threonine phosphorylated:
T447, T458, T1271, T1493+, T1494+, T1505+.
Tyrosine phosphorylated:
Y81, Y138, Y140, Y442, Y582, Y1076, Y1548, Y1556.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 29
29
865
29
921
 2
2
63
17
27
 3
3
104
53
76
 12
12
374
135
466
 34
34
1027
25
728
 19
19
561
82
1372
 13
13
378
35
498
 11
11
318
84
600
 25
25
740
17
550
 4
4
118
138
72
 3
3
85
75
83
 20
20
607
237
702
 5
5
159
75
116
 3
3
98
15
81
 4
4
128
70
121
 4
4
129
16
68
 4
4
109
246
80
 4
4
118
64
134
 6
6
171
134
100
 28
28
832
109
746
 3
3
96
68
73
 4
4
128
71
117
 4
4
129
63
102
 5
5
156
64
135
 5
5
145
68
143
 15
15
459
100
646
 5
5
159
78
109
 4
4
117
64
98
 5
5
156
64
109
 1
1
30
28
11
 29
29
874
36
700
 100
100
2996
36
5019
 27
27
798
68
1399
 35
35
1054
57
852
 7
7
196
44
234
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
97 -
-
-
99 -
-
-
92 -
-
-
- 93
93
95.5
94 -
-
-
- 88.3
88.3
92.4
90 -
-
-
91 -
-
-
- 38.9
38.9
41.8
- 84.5
84.5
90.6
86.5 -
-
-
77 67
67
78.2
73 -
-
-
- 25.2
25.2
42
37 29.6
29.6
47.1
- -
-
-
- 36.1
36.1
53
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by phosphorylation on Thr-1505. ; The GADD45 proteins activate the kinase by binding to the N-terminal domain.
Inhibition:
N-terminal autoinhibitory domain interacts with the C-terminal kinase domain, inhibiting kinase activity, and preventing interaction with its substrate, MAP2K6.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MEKK4 (MAP3K4) | Q9Y6R4 | T1493 | KLKNNAQTMPGEVNS | + |
| MKK4 (MAP2K4, MEK4) | P45985 | S257 | ISGQLVDSIAKTRDA | + |
| MKK4 (MAP2K4, MEK4) | P45985 | T261 | LVDSIAKTRDAGCRP | + |
| MKK6 (MAP2K6, MEK6) | P52564 | S207 | ISGYLVDSVAKTIDA | + |
| MKK6 (MAP2K6, MEK6) | P52564 | T211 | LVDSVAKTIDAGCKP | + |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
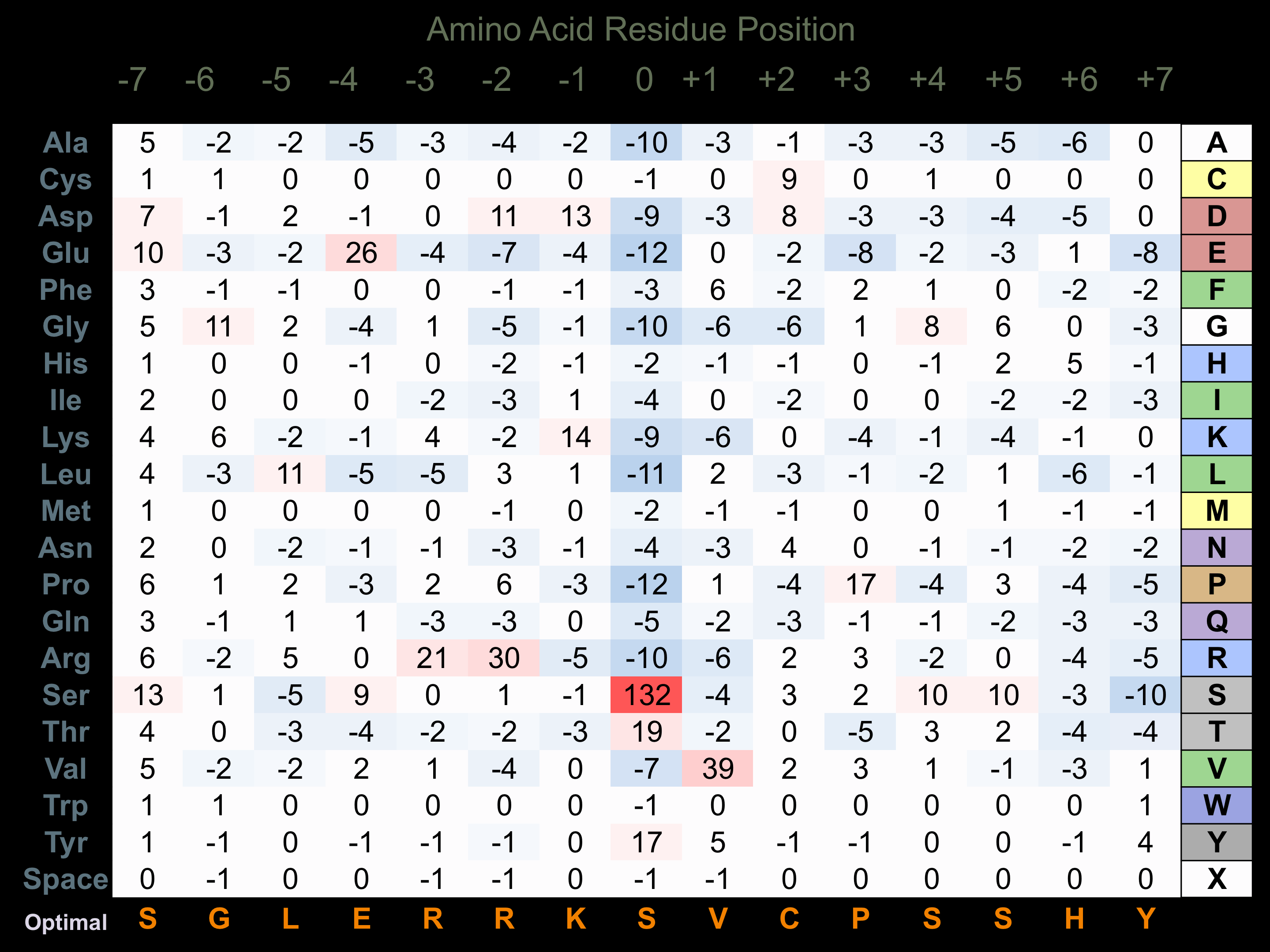
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Hesperadin | Kd < 10 nM | 10142586 | 514409 | 19035792 |
| Neratinib | Kd = 39 nM | 9915743 | 180022 | 22037378 |
| PD173955 | Kd = 56 nM | 447077 | 386051 | 22037378 |
| Bosutinib | Kd = 110 nM | 5328940 | 288441 | 22037378 |
| Pelitinib | Kd = 130 nM | 6445562 | 607707 | 15711537 |
| Dasatinib | Kd = 310 nM | 11153014 | 1421 | 18183025 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| Staurosporine | Kd = 1.2 µM | 5279 | 18183025 | |
| KW2449 | Kd = 1.6 µM | 11427553 | 1908397 | 22037378 |
| PHA-665752 | Kd = 1.8 µM | 10461815 | 450786 | 22037378 |
| Foretinib | Kd = 1.9 µM | 42642645 | 1230609 | 22037378 |
| Canertinib | Kd = 2 µM | 156414 | 31965 | 15711537 |
| NVP-TAE684 | Kd = 2 µM | 16038120 | 509032 | 22037378 |
| Tozasertib | Kd = 2.5 µM | 5494449 | 572878 | 18183025 |
| Lestaurtinib | Kd = 2.6 µM | 126565 | 22037378 | |
| Vandetanib | Kd = 3 µM | 3081361 | 24828 | 22037378 |
| Sunitinib | Kd = 3.3 µM | 5329102 | 535 | 15711537 |
| TG101348 | Kd = 4.1 µM | 16722836 | 1287853 | 22037378 |
Disease Linkage
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +59, p<0.003); Brain glioblastomas (%CFC= +796, p<0.103); Breast epithelial cell carcinomas (%CFC= +45, p<(0.0003); Cervical cancer stage 2A (%CFC= +69, p<0.048); Prostate cancer (%CFC= +45, p<0.032); Skin melanomas (%CFC= -51, p<0.098); and Skin melanomas - malignant (%CFC= -76, p<0.0001). The COSMIC website notes an up-regulated expression score for MEKK4 in diverse human cancers of 355, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 139 for this protein kinase in human cancers was 2.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A K1372R mutation can abrogate phosphotransferase activity of MEKK4 and inhibit activation of the p38 and JNK MAPK pathways.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25012 diverse cancer specimens. This rate is only -1 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.49 % in 1093 large intestine cancers tested; 0.22 % in 805 skin cancers tested; 0.2 % in 602 endometrium cancers tested; 0.2 % in 589 stomach cancers tested; 0.13 % in 1808 lung cancers tested; 0.1 % in 830 upper aerodigestive tract cancers tested; 0.09 % in 1292 breast cancers tested.
Frequency of Mutated Sites:
None > 6 in 20,224 cancer specimens
Comments:
Over 26 deletions (16 at A1199delA, 6 at N160fs*8), 6 insertions, and no complex mutations are noted on the COSMIC website.

