Nomenclature
Short Name:
MISR2
Full Name:
Anti-Muellerian hormone type II receptor
Alias:
- AMH type II receptor
- MIS type II receptor
- MISRII
- MRII
- AMHR2
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
STKR
SubFamily:
Type2
Structure
Mol. Mass (Da):
62,750
# Amino Acids:
573
# mRNA Isoforms:
3
mRNA Isoforms:
62,750 Da (573 AA; Q16671); 52,241 Da (478 AA; Q16671-2); 51,827 Da (478 AA; Q16671-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
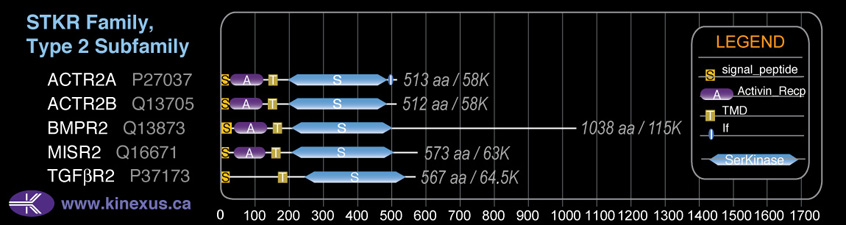
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 17 | signal_peptide |
| 38 | 124 | Activin_recp |
| 148 | 170 | TMD |
| 203 | 513 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
N-GlcNAcylated:
N66, N119.
Serine phosphorylated:
S189.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 8
8
195
16
220
 3
3
64
10
12
 2
2
43
11
44
 22
22
534
53
1867
 10
10
232
14
233
 83
83
1996
45
4614
 11
11
256
19
503
 33
33
795
28
1765
 6
6
152
10
155
 2
2
48
52
71
 2
2
45
22
73
 21
21
502
113
633
 1
1
34
22
77
 1
1
24
8
17
 3
3
71
19
97
 5
5
113
7
83
 0.7
0.7
16
106
54
 6
6
155
16
454
 1
1
29
53
26
 9
9
208
56
288
 2
2
39
18
52
 3
3
62
21
82
 2
2
39
20
49
 3
3
65
17
136
 4
4
93
18
258
 58
58
1380
36
3024
 0.8
0.8
20
25
33
 3
3
63
16
97
 2
2
43
16
63
 2
2
57
14
18
 18
18
422
18
307
 100
100
2397
21
4290
 16
16
391
38
952
 30
30
726
31
757
 0.9
0.9
22
22
23
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.6
99.6
99.8
100 97.2
97.2
98.2
97 -
-
-
84.5 -
-
-
89 82.1
82.1
86.5
86 -
-
-
- 78.5
78.5
86
79 77.4
77.4
84.4
79 -
-
-
- -
-
-
- 28.1
28.1
45.7
- 27.5
27.5
45.2
40.5 -
-
-
- -
-
-
- 21
21
32.1
- 28.7
28.7
45.2
- 23
23
39.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | AMH - P03971 |
| 2 | BMPR1B - O00238 |
| 3 | TGFBR1 - P36897 |
| 4 | PTEN - P60484 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
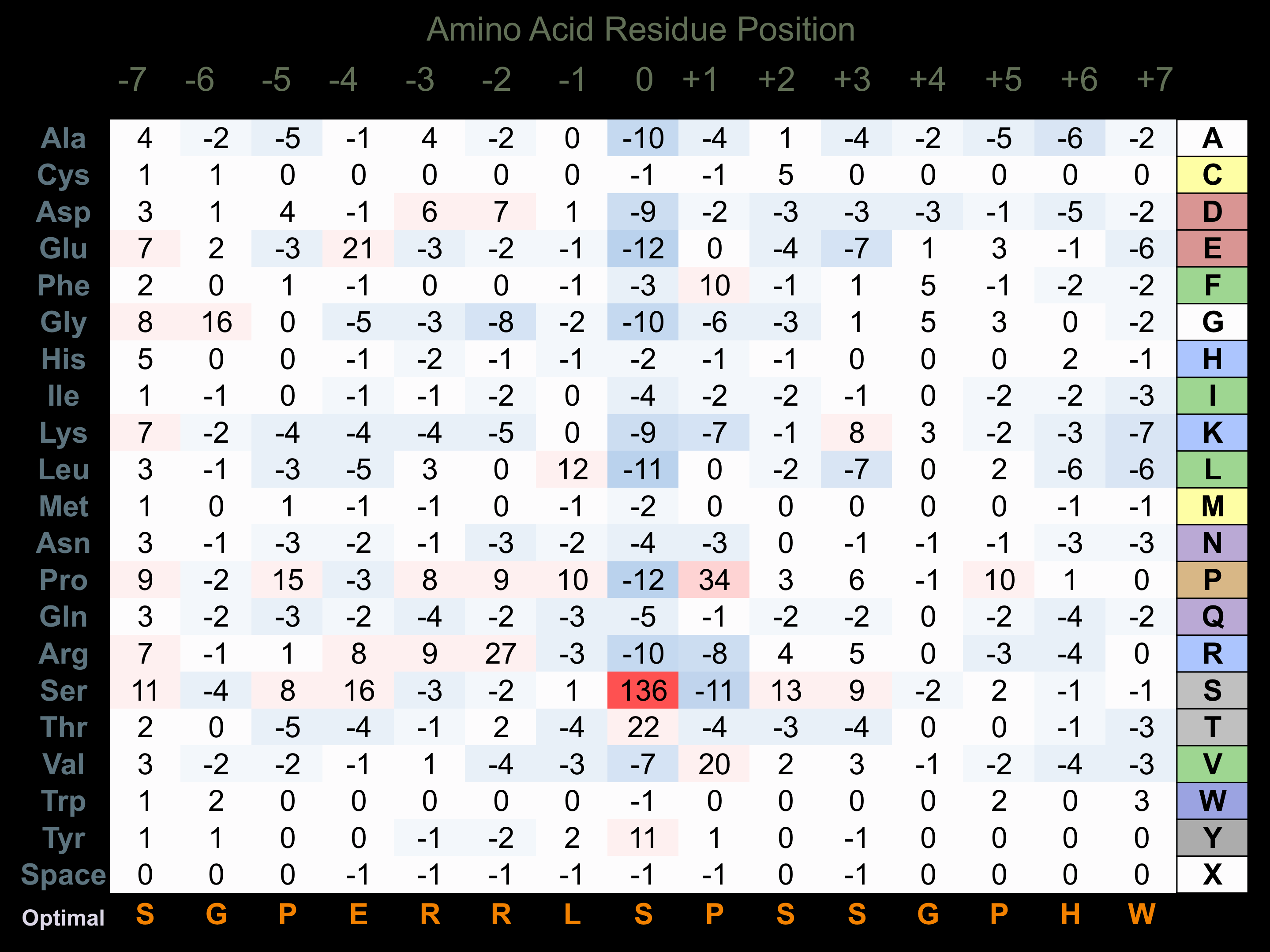
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer, endocrine disorders
Specific Diseases (Non-cancerous):
Persistent Mullerian duct syndrome, Type 1; Persistent Mullerian duct syndrome; Persistent Mullerian duct syndrome, Type 1I; Mixed gonadal dysgenesis
Comments:
Mutations of MISR2 at 54, 142, 282, 406, 426 and other amino acid positions are associated with persistent Mullerian duct syndrome 2, which is a form of male pseudohermaphroditism due to failure of Muellerian duct regression.
Specific Cancer Types:
Ovarian cancer
Comments:
MISR2 variants are associated with an increased risk of breast cancer. Homozygous A482G substitution is associated with higher risks of polycistic ovary syndrome. Women with polycistic ovary syndrome may have enlarged ovaries with small collections of follicles, and tend to have infrequent or prolonged menstrual periods.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for MISR2 in diverse human cancers of 307, which is 0.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 0 for this protein kinase in human cancers was 100% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24998 diverse cancer specimens. This rate is only 16 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.69 % in 864 skin cancers tested; 0.27 % in 1270 large intestine cancers tested; 0.26 % in 603 endometrium cancers tested; 0.19 % in 1634 lung cancers tested; 0.15 % in 589 stomach cancers tested; 0.15 % in 238 bone cancers tested; 0.14 % in 127 biliary tract cancers tested; 0.13 % in 273 cervix cancers tested; 0.1 % in 710 oesophagus cancers tested; 0.07 % in 1316 breast cancers tested; 0.06 % in 942 upper aerodigestive tract cancers tested; 0.06 % in 833 ovary cancers tested; 0.06 % in 558 thyroid cancers tested; 0.04 % in 441 autonomic ganglia cancers tested; 0.04 % in 1459 pancreas cancers tested; 0.04 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: E248K (4).
Comments:
Only 2 deletions, 1 insertion, and no complex mutations are noted on the COSMIC website.

