Nomenclature
Short Name:
MLK2
Full Name:
Mitogen-activated protein kinase kinase kinase 10
Alias:
- EC 2.7.11.25
- Kinase MLK2
- MKN28 derived nonreceptor_type serine/threonine kinase
- MKN28 kinase
- MST
- Protein kinase MST
- M3K10
- MAP3K10
- MEKK10
- Mitogen-activated protein kinase kinase kinase 10
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
MLK
SubFamily:
MLK
Specific Links
Structure
Mol. Mass (Da):
103694
# Amino Acids:
954
# mRNA Isoforms:
1
mRNA Isoforms:
103,694 Da (954 AA; Q02779)
4D Structure:
Homodimer
1D Structure:
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 16 | 81 | SH3 |
| 98 | 360 | Pkinase |
| 378 | 449 | Coiled-coil |
| 384 | 405 | leucine zipper 1 |
| 419 | 440 | Leucine zipper 2 |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S262+, S446, S489, S498, S502, S506, S508, S589, S809, S830+.
Threonine phosphorylated:
T258+, T266-, T558.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 10
10
333
19
487
 0.7
0.7
22
10
14
 3
3
87
10
107
 30
30
962
63
2325
 19
19
602
13
572
 3
3
93
44
261
 9
9
297
19
578
 43
43
1366
38
3009
 11
11
355
10
363
 3
3
85
55
138
 2
2
55
24
62
 19
19
593
106
732
 1.2
1.2
38
21
64
 0.4
0.4
12
6
1
 2
2
61
21
81
 0.3
0.3
10
8
9
 7
7
227
191
2693
 2
2
69
16
82
 1.2
1.2
39
59
44
 10
10
310
56
381
 2
2
59
20
59
 1.2
1.2
38
22
50
 2
2
57
27
51
 2
2
51
17
74
 2
2
74
19
92
 25
25
800
44
1483
 1
1
31
24
45
 2
2
49
15
73
 1.3
1.3
43
17
57
 2
2
72
14
11
 23
23
721
18
522
 100
100
3203
21
4034
 6
6
205
41
484
 19
19
619
36
710
 1
1
33
22
27
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 52.3
52.3
63.7
96 94.3
94.3
95.8
- -
-
-
95 -
-
-
- 46.5
46.5
57.2
93 -
-
-
- 92.1
92.1
94.3
92.5 46.7
46.7
58
93 -
-
-
- -
-
-
- -
-
-
- 60.9
60.9
71
68 59.2
59.2
68.7
68 -
-
-
- 31.8
31.8
45.3
- 34.2
34.2
48.6
- 20.3
20.3
35.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
39 -
-
-
38 -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PHB - P35232 |
| 2 | MAP2K4 - P45985 |
| 3 | TUBB2A - Q13885 |
| 4 | DNM1 - Q05193 |
| 5 | DLG4 - P78352 |
| 6 | CLTC - Q00610 |
| 7 | DNM1L - O00429 |
| 8 | CEACAM1 - P13688 |
| 9 | SH3RF1 - Q7Z6J0 |
| 10 | NEUROD1 - Q13562 |
| 11 | CNKSR1 - Q969H4 |
| 12 | MAPK8IP2 - Q13387 |
| 13 | MAPK8IP1 - Q9UQF2 |
| 14 | HGS - O14964 |
| 15 | CDC42 - P60953 |
Regulation
Activation:
Homodimerization via the leucine zipper domains is required for autophosphorylation and subsequent activation. Autophosphorylation on serine and threonine residues within the activation loop plays a role in enzyme activation.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
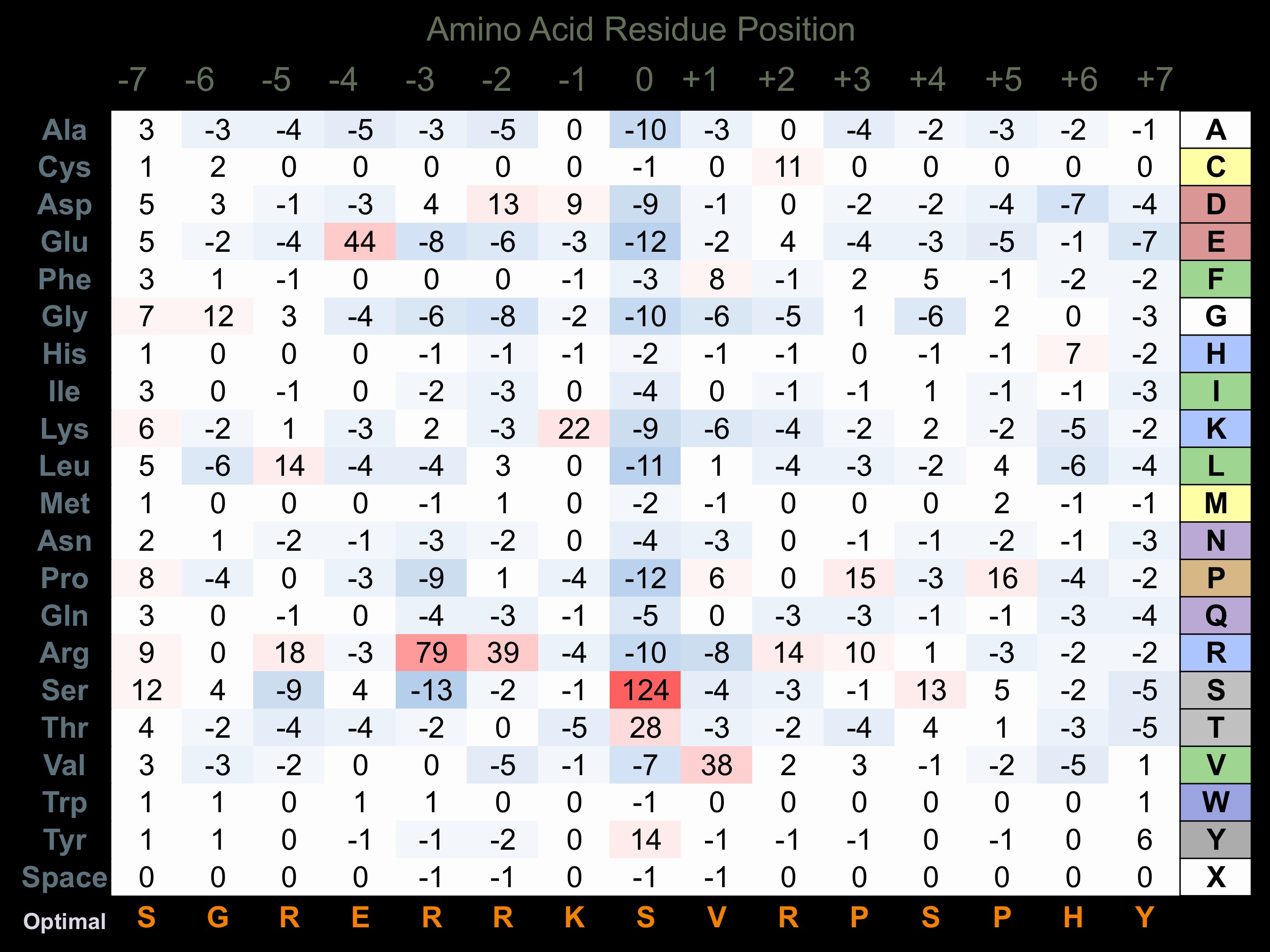
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Neuroblastomas (NB); Differentiating neuroblastomas
Comments:
MLK2 promotes the proliferation and decreases the sensitivity of pancreatic cancer cells to gemcitabine.
Gene Expression in Cancers:
The COSMIC website notes an up-regulated expression score for MLK2 in diverse human cancers of 673, which is 1.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 38 for this protein kinase in human cancers was 0.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25371 diverse cancer specimens. This rate is only -15 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.32 % in 1093 large intestine cancers tested; 0.29 % in 805 skin cancers tested; 0.24 % in 830 upper aerodigestive tract cancers tested; 0.21 % in 602 endometrium cancers tested; 0.06 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: Q88P (15); Q91P (14).
Comments:
Only 6 deletions, 1 insertion and no complex mutations are noted on the COSMIC website.

