Nomenclature
Short Name:
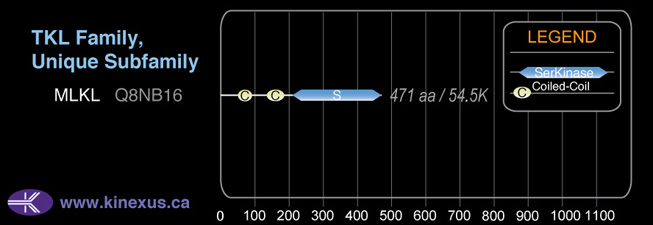
MLKL
Full Name:
MLKL protein
Alias:
- Mixed lineage kinase domain-like protein
Classification
Type:
Protein-serine/threonine kinase
Group:
TKL
Family:
TKL-Unique
SubFamily:
NA
Structure
Mol. Mass (Da):
54,479
# Amino Acids:
471
# mRNA Isoforms:
2
mRNA Isoforms:
54,479 Da (471 AA; Q8NB16); 30,279 Da (263 AA; Q8NB16-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 55 | 82 | Coiled-coil |
| 148 | 175 | Coiled-coil |
| 203 | 468 | Pkinase |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K354.
Serine phosphorylated:
S52, S92, S125, S128, S161, S334, S358, S373, S393, S417.
Threonine phosphorylated:
T59, T246, T302, T357, T364, T374.
Ubiquitinated:
K331.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 57
57
1599
9
1287
 28
28
789
9
1546
 -
-
-
-
-
 6
6
178
33
481
 20
20
565
16
599
 4
4
111
9
21
 3
3
87
16
36
 14
14
381
10
841
 2
2
47
6
29
 13
13
360
34
978
 17
17
488
10
976
 33
33
937
16
778
 100
100
2810
2
527
 21
21
582
8
1391
 16
16
444
10
990
 10
10
280
7
441
 3
3
81
73
194
 17
17
477
8
930
 13
13
369
26
1020
 35
35
983
36
1021
 14
14
380
10
737
 7
7
203
8
197
 -
-
-
-
-
 42
42
1186
10
2135
 13
13
377
10
598
 26
26
725
24
609
 18
18
519
8
854
 17
17
472
8
919
 14
14
404
8
877
 9
9
252
14
125
 62
62
1750
12
44
 18
18
501
13
871
 19
19
527
12
172
 49
49
1364
26
847
 4
4
117
13
34
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 97.7
97.7
99.6
98 90.5
90.5
95.5
91 -
-
-
65 -
-
-
- -
-
-
- -
-
-
- 61.9
61.9
77.5
64 62
62
77.5
64 -
-
-
- 45
45
61.8
- -
-
-
41.5 -
-
-
38 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20.6
20.6
34.7
- 23.1
23.1
43.1
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PITPNA - Q00169 |
| 2 | PBX2 - P40425 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
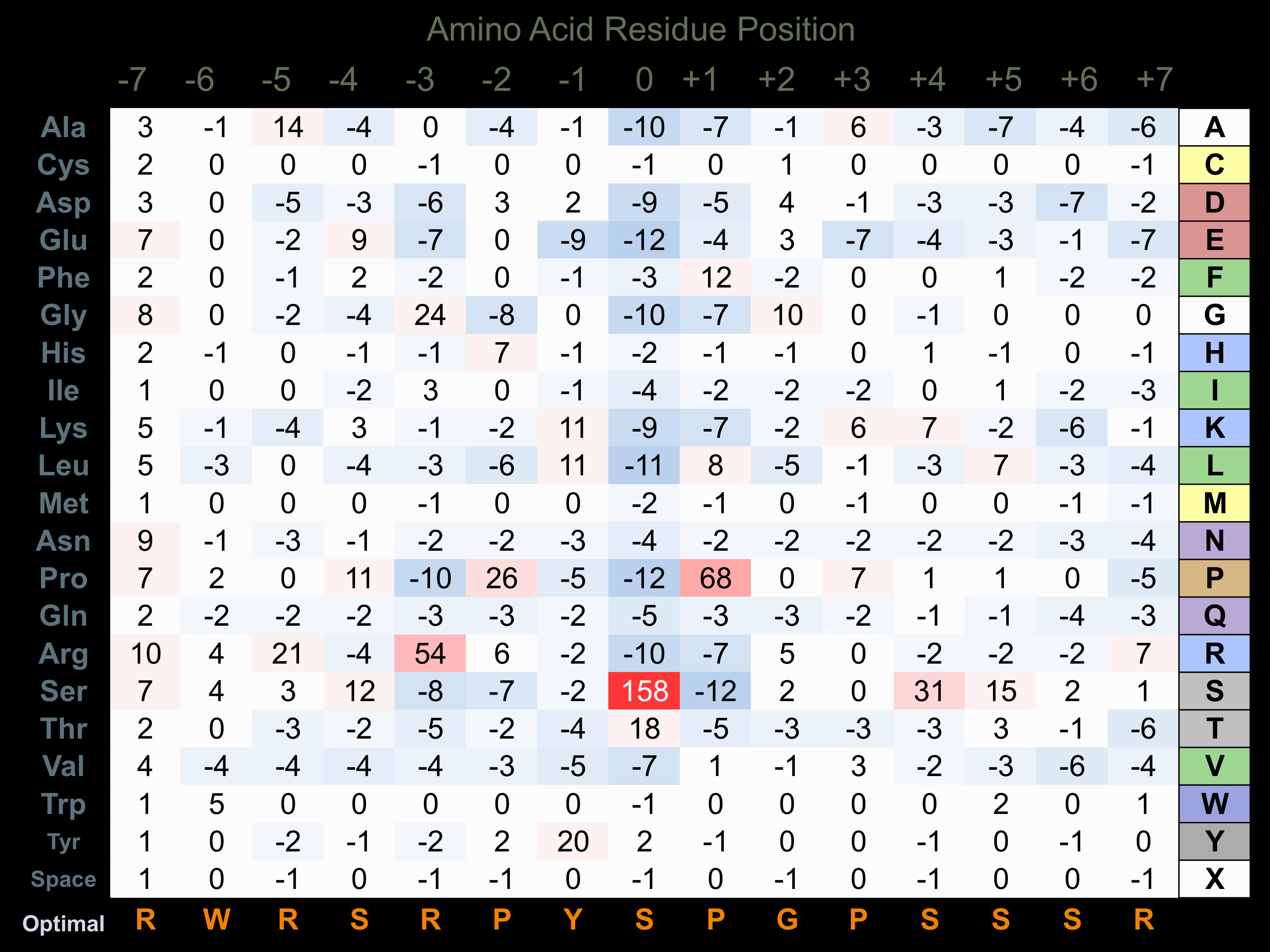
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
Comments:
MLKL plays a role in TNF-induced necroptosis, a regulated form of necrosis. It is activated by phosphorylation, leads to homotrimerization, and executes programmed necrosis by compromising plasma membrane integrity. Mutations at multiple sites are associated with abolished or impaired ability to mediate necroptosis.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Clear cell renal cell carcinomas (cRCC) (%CFC= +74, p<0.014); Colon mucosal cell adenomas (%CFC= +53, p<0.0001); and Ovary adenocarcinomas (%CFC= +112, p<0.005). The COSMIC website notes an up-regulated expression score for MLKL in diverse human cancers of 279, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 26 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 24954 diverse cancer specimens. This rate is very similar (+ 10% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.52 % in 864 skin cancers tested; 0.42 % in 1270 large intestine cancers tested; 0.32 % in 603 endometrium cancers tested; 0.3 % in 629 stomach cancers tested; 0.16 % in 273 cervix cancers tested; 0.1 % in 1822 lung cancers tested; 0.09 % in 238 bone cancers tested; 0.08 % in 1276 kidney cancers tested; 0.06 % in 382 soft tissue cancers tested; 0.06 % in 1512 liver cancers tested; 0.04 % in 548 urinary tract cancers tested; 0.04 % in 2009 haematopoietic and lymphoid cancers tested; 0.03 % in 833 ovary cancers tested; 0.03 % in 710 oesophagus cancers tested; 0.03 % in 1316 breast cancers tested; 0.02 % in 942 upper aerodigestive tract cancers tested; 0.02 % in 881 prostate cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,237 cancer specimens
Comments:
Only 5 deletions, no insertions and 1 complex mutation noted on the COSMIC website.

