Nomenclature
Short Name:
MOK
Full Name:
MAPK-MAK-MRK overlapping kinase
Alias:
- EC 2.7.11.22
- RAGE
- RAGE1
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
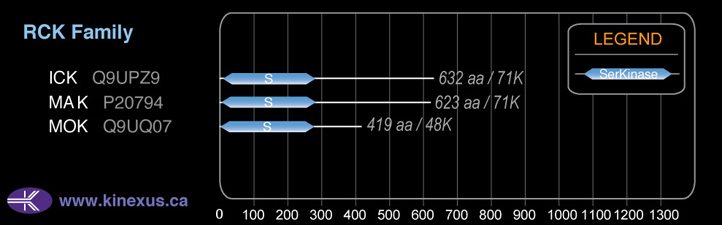
Family:
RCK
SubFamily:
NA
Structure
Mol. Mass (Da):
48,014
# Amino Acids:
419
# mRNA Isoforms:
6
mRNA Isoforms:
48,014 Da (419 AA; Q9UQ07); 47,886 Da (418 AA; Q9UQ07-6); 44,425 Da (389 AA; Q9UQ07-5); 27,120 Da (231 AA; Q9UQ07-2); 10,482 Da (95 AA; Q9UQ07-4); 7,879 Da (73 AA; Q9UQ07-3)
4D Structure:
NA
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 4 | 285 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S75, S77, S298, S330, S391, S399.
Threonine phosphorylated:
T159+, T220.
Tyrosine phosphorylated:
Y29, Y161+.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 25
25
739
19
1062
 3
3
85
12
69
 2
2
50
12
57
 48
48
1406
71
2572
 25
25
728
14
678
 5
5
140
48
262
 17
17
499
23
680
 65
65
1925
44
3207
 7
7
211
10
191
 6
6
176
70
158
 2
2
72
29
112
 14
14
408
138
575
 2
2
45
23
37
 3
3
80
12
51
 5
5
141
15
179
 3
3
92
9
63
 6
6
176
197
1664
 3
3
93
19
118
 2
2
48
60
33
 15
15
445
56
469
 3
3
94
25
74
 2
2
47
25
45
 2
2
67
21
63
 14
14
424
21
400
 2
2
53
25
50
 100
100
2954
50
4490
 1
1
29
26
24
 5
5
149
21
114
 3
3
88
21
75
 14
14
403
14
125
 26
26
770
18
551
 28
28
818
21
1416
 8
8
237
49
512
 21
21
608
31
620
 42
42
1240
22
2093
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.5
96 82.7
82.7
85.3
- -
-
-
82 -
-
-
58 76.6
76.6
81.2
85 -
-
-
- 83.6
83.6
89.5
84 27.8
27.8
42.5
96 -
-
-
- -
-
-
- 28.6
28.6
43.9
65 63.3
63.3
73.3
67 -
-
-
- -
-
-
- 26.7
26.7
42.7
- -
-
-
- -
-
-
- 50.7
50.7
68.4
- -
-
-
- -
-
-
- -
-
-
- 30.5
30.5
47.6
- 28.6
28.6
43.2
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Phosphorylation appears to increase the enzymatic activity
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
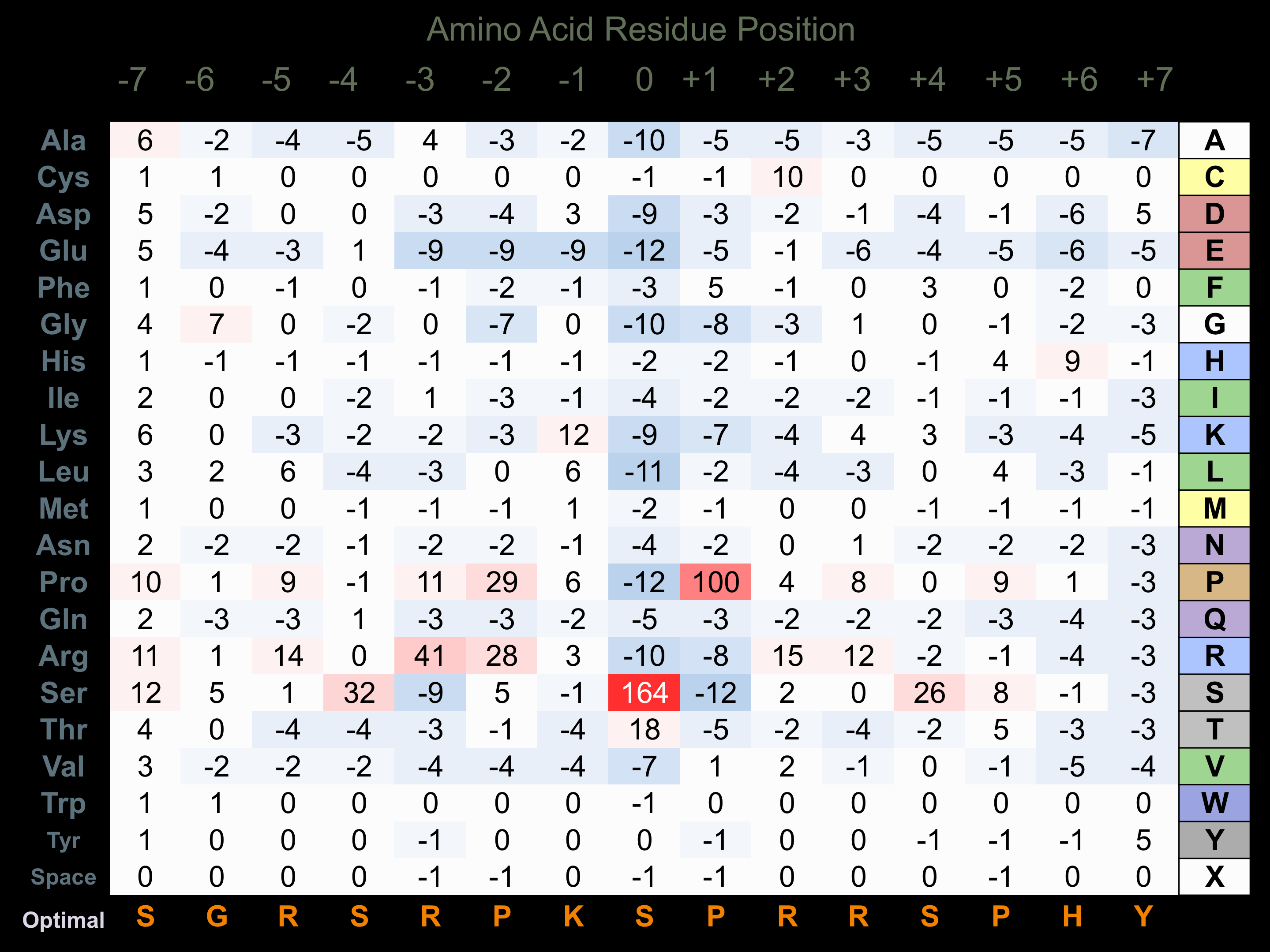
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Renal cell carcinomas (RCC)
Comments:
MOK may be a tumour requiring protein (TRP), since it displays extremely low rates of mutation in human cancers. MOK is a renal tumour antigen that is silent in most normal tissues except for retina and a number of tumour cell types.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Large B-cell lymphomas (%CFC= +59, p<(0.0003); Pituitary adenomas (ACTH-secreting) (%CFC= +68); and Skin melanomas - malignant (%CFC= +229, p<0.0001).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.01 % in 24433 diverse cancer specimens. This rate is -87 % lower than the average rate of 0.075 % calculated for human protein kinases in general. Such a very low frequency of mutation in human cancers is consistent with this protein kinase playing a role as a tumour requiring protein (TRP).
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 603 endometrium cancers tested; 0.06 % in 807 ovary cancers tested; 0.06 % in 1253 kidney cancers tested;
Frequency of Mutated Sites:
None > 2 in 19,689 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.

