Nomenclature
Short Name:
MRCKa
Full Name:
Myotonic dystrophy kinase-related CDC42-binding kinase alpha
Alias:
- CDC42BPA
- CDC42-binding protein kinase alpha
- KIAA0451
- MRCK alpha
- PK428
- DMPK-like alpha
- DKFZp686L1738
- EC 2.7.11.1
- FLJ23347
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
DMPK
SubFamily:
GEK
Structure
Mol. Mass (Da):
197307
# Amino Acids:
1732
# mRNA Isoforms:
6
mRNA Isoforms:
202,812 Da (1781 AA; Q5VT25-6); 199,811 Da (1754 AA; Q5VT25-2); 197,307 Da (1732 AA; Q5VT25); 195,922 Da (1719 AA; Q5VT25-5); 193,031 Da (1691 AA; Q5VT25-4); 186,113 Da (1638 AA; Q5VT25-3)
4D Structure:
Homodimer and homotetramer via the coiled coil regions. Interacts tightly with GTP-bound but not GDP-bound CDC42.
1D Structure:
Subfamily Alignment
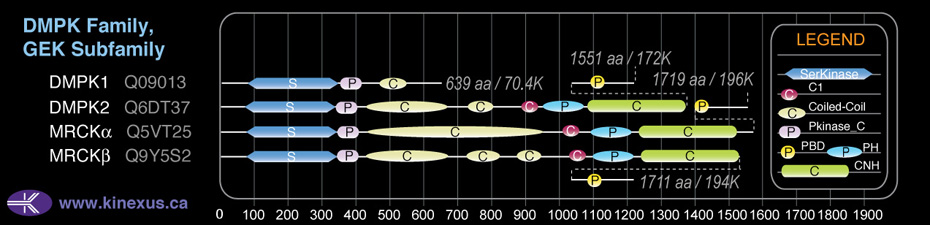
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 77 | 343 | Pkinase |
| 344 | 414 | Pkinase_C |
| 437 | 820 | Coiled-coil |
| 880 | 943 | Coiled-coil |
| 1012 | 1062 | C1 |
| 1082 | 1201 | PH |
| 1227 | 1499 | CNH |
| 1571 | 1584 | PBD |
| 1571 | 1584 | CRIB |
| 881 | 941 | DMPK coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K669.
Serine phosphorylated:
S2, S222, S234+, S475, S674, S725, S843, S856, S1127, S1171, S1173, S1378, S1545, S1604, S1611, S1613, S1616, S1629, S1632, S1635, S1639, S1651, S1654, S1656, S1664, S1669, S1672, S1693, S1700, S1703, S1706, S1710, S1719, S1721, S1724, S1729.
Threonine phosphorylated:
T240+, T403+, T846, T862, T992, T994, T1185, T1505, T1618, T1725.
Tyrosine phosphorylated:
Y444, Y474, Y1510, Y1544, Y1655.
Ubiquitinated:
K338, K1122.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 90
90
873
64
955
 7
7
67
28
75
 67
67
648
16
1066
 40
40
388
200
560
 84
84
813
55
640
 7
7
69
165
158
 22
22
211
69
398
 98
98
943
71
1620
 77
77
740
31
613
 16
16
156
180
215
 13
13
129
52
170
 75
75
724
322
667
 5
5
50
60
84
 12
12
118
21
133
 16
16
158
33
266
 8
8
82
35
104
 16
16
152
293
553
 13
13
126
30
198
 8
8
80
181
114
 54
54
524
243
572
 22
22
213
37
298
 11
11
102
44
152
 36
36
350
19
445
 34
34
330
31
645
 5
5
47
37
65
 100
100
965
122
1734
 5
5
49
62
91
 13
13
123
31
167
 24
24
233
30
423
 21
21
207
70
244
 30
30
292
48
287
 68
68
655
77
1282
 95
95
913
158
1633
 82
82
795
130
696
 46
46
447
83
460
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 59.6
59.6
76.3
91 59.6
59.6
76.3
99 -
-
-
96 -
-
-
- 95.7
95.7
96.3
96 96.9
96.9
99.1
- -
-
-
96 95.4
95.4
97.8
96 95.9
95.9
98.3
- 88.4
88.4
94.5
- 88.4
88.4
94.5
88 -
-
-
80.5 73.3
73.3
86.3
74 -
-
-
- -
-
-
43 44.3
44.3
63.7
- -
-
-
45 24.5
24.5
42.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MYL9 - P24844 |
| 2 | MYL12A - P19105 |
| 3 | MYL2 - P10916 |
| 4 | CDC42 - P60953 |
| 5 | RHOQ - P17081 |
| 6 | PPP1R12C - Q9BZL4 |
Regulation
Activation:
Agonist binding to the phorbol ester binding site disrupts this, releasing the kinase domain to allow N-terminus-mediated dimerization and kinase activation by transautophosphorylation.
Inhibition:
Maintained in an inactive, closed conformation by an interaction between the kinase domain and the negative autoregulatory C-terminal coiled-coil region.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
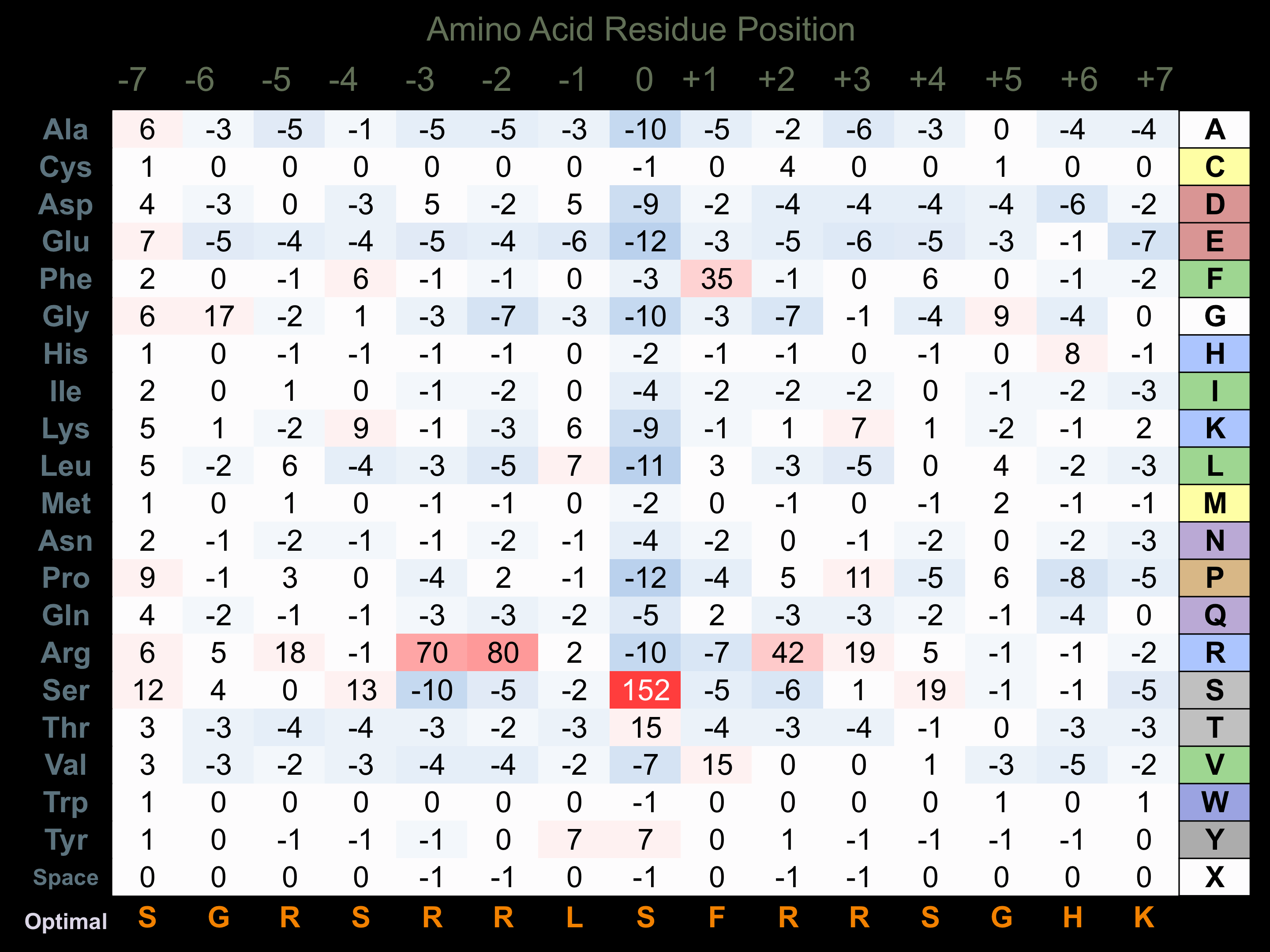
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| Staurosporine | Kd = 57 nM | 5279 | 18183025 | |
| AT9283 | IC50 > 100 nM | 24905142 | 19143567 | |
| A674563 | Kd = 380 nM | 11314340 | 379218 | 22037378 |
| Cdk1/2 Inhibitor III | IC50 = 500 nM | 5330812 | 261720 | 22037377 |
| Foretinib | Kd = 660 nM | 42642645 | 1230609 | 22037378 |
| A 443654 | IC50 < 1 µM | 10172943 | 379300 | 19465931 |
| Kinome_714 | IC50 > 1 µM | 46886323 | 20346655 | |
| MK5108 | IC50 > 1 µM | 24748204 | 20053775 | |
| Silmitasertib | IC50 > 1 µM | 24748573 | 21174434 | |
| SNS314 | IC50 > 1 µM | 16047143 | 514582 | 18678489 |
| SU11652 | IC50 > 1 µM | 24906267 | 13485 | 22037377 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| CHEMBL573339 | Kd = 1.8 µM | 9884685 | 573339 | 18183025 |
| PI-103 | Kd = 1.8 µM | 16739368 | 538346 | 18183025 |
| Dasatinib | Kd = 2 µM | 11153014 | 1421 | 18183025 |
| Momelotinib | IC50 > 2 µM | 25062766 | 19295546 | |
| LKB1(AAK1 dual inhibitor) | Kd < 2.5 µM | 44588117 | 516312 | 19035792 |
| Vandetanib | Kd = 2.6 µM | 3081361 | 24828 | 18183025 |
| JNJ-28871063 | IC50 > 4 µM | 17747413 | 17975007 | |
| Kinome_2917 | Ki = 4.44 µM | 11362471 | 223367 | 17352464 |
| BMS-690514 | Kd < 4.5 µM | 11349170 | 21531814 |
Disease Linkage
General Disease Association:
Musculoskeletal disorders
Specific Diseases (Non-cancerous):
Myotonic dystrophy
Comments:
By FISH, the MRCK-a gene is identified to show association with rippling muscle disease (RMD), which is characterized by mechanically triggered contractions of skeletal muscle.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +125, p<0.002); Cervical cancer (%CFC= -59, p<0.0001); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -79, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +95, p<0.0001); and Prostate cancer - primary (%CFC= -80, p<0.0001).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 24433 diverse cancer specimens. This rate is -43 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.25 % in 1270 large intestine cancers tested; 0.19 % in 603 endometrium cancers tested; 0.18 % in 548 urinary tract cancers tested; 0.16 % in 864 skin cancers tested; 0.12 % in 1823 lung cancers tested; 0.11 % in 589 stomach cancers tested; 0.08 % in 273 cervix cancers tested; 0.08 % in 1512 liver cancers tested; 0.08 % in 1316 breast cancers tested; 0.07 % in 238 bone cancers tested; 0.05 % in 710 oesophagus cancers tested; 0.05 % in 127 biliary tract cancers tested; 0.04 % in 1276 kidney cancers tested; 0.03 % in 833 ovary cancers tested; 0.03 % in 1459 pancreas cancers tested; 0.02 % in 942 upper aerodigestive tract cancers tested; 0.02 % in 382 soft tissue cancers tested; 0.01 % in 881 prostate cancers tested; 0.01 % in 558 thyroid cancers tested; 0.01 % in 2082 central nervous system cancers tested; 0.01 % in 2009 haematopoietic and lymphoid cancers tested.
Frequency of Mutated Sites:
None > 3 in 19,689 cancer specimens
Comments:
Only 5 deletions, 5 insertions and 2 complex mutations noted on the COSMIC website.

