Nomenclature
Short Name:
NDR1
Full Name:
Serine-threonine-protein kinase 38
Alias:
- AA617404
- DJ108K11.2
- EC 2.7.11.1
- NDR1 protein kinase
- STK38
Classification
Type:
Protein-serine/threonine kinase
Group:
AGC
Family:
NDR
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
54190
# Amino Acids:
465
# mRNA Isoforms:
1
mRNA Isoforms:
54,190 Da (465 AA; Q15208)
4D Structure:
Homodimeric S100B binds two molecules of STK38. Component of the MLL5-L complex, at least composed of MLL5, STK38, PPP1CA, PPP1CB, PPP1CC, HCFC1, ACTB and OGT. Interacts with MOB1 and MOB2. Interacts with MAP3K1 and MAP3K2 (via the kinase catalytic domain).
1D Structure:
Subfamily Alignment
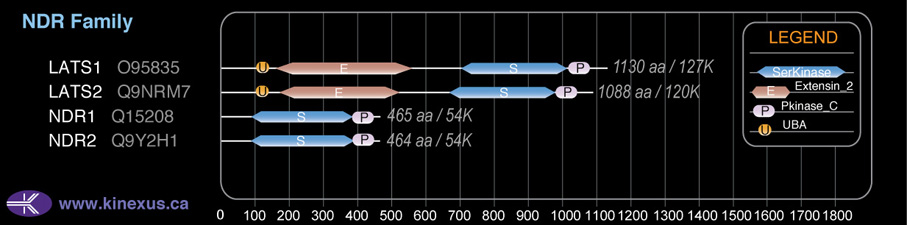
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
A2.
Serine phosphorylated:
S6, S10, S11, S13, S32, S264, S281+.
Threonine phosphorylated:
T4, T7, T16, T26, T74, T183, T235, T270, T282, T285-, T407, T444+, T452.
Tyrosine phosphorylated:
Y31, Y288, Y443, Y445, Y460.
Ubiquitinated:
K62, K93, K214.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 39
39
993
54
942
 1.1
1.1
29
25
27
 16
16
400
15
350
 11
11
281
186
731
 33
33
859
45
597
 3
3
83
132
126
 15
15
375
71
535
 21
21
542
64
810
 23
23
580
24
604
 4
4
111
145
142
 5
5
117
50
151
 25
25
648
285
609
 9
9
222
48
333
 2
2
45
21
42
 3
3
89
31
112
 1.1
1.1
29
34
35
 5
5
140
318
915
 5
5
136
30
173
 3
3
67
135
75
 24
24
623
208
643
 3
3
89
40
102
 10
10
256
45
339
 9
9
236
26
267
 5
5
140
30
171
 12
12
314
40
428
 26
26
677
114
819
 7
7
185
51
260
 7
7
189
30
258
 8
8
195
30
248
 3
3
82
42
49
 45
45
1160
30
1063
 100
100
2573
59
7602
 9
9
235
110
411
 33
33
844
135
707
 29
29
738
74
2565
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.8
100 99.8
99.8
99.8
100 -
-
-
99 -
-
-
- 98.5
98.5
98.7
100 76.2
76.2
76.2
- -
-
-
99 98.7
98.7
99.3
98 -
-
-
- -
-
-
- 87.5
87.5
88.5
98 87.5
87.5
88.5
93 93.1
93.1
96.1
92 91.7
91.7
95.1
- -
-
-
73 68.6
68.6
83.7
- -
-
-
66 63.2
63.2
75
- -
-
-
- -
-
-
- -
-
-
- -
-
-
48 -
-
-
- 33.3
33.3
46.2
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | MOBKL1B - Q9H8S9 |
| 2 | MOB2 - Q70IA6 |
| 3 | S100B - P04271 |
| 4 | CTDP1 - Q9Y5B0 |
| 5 | PRMT5 - O14744 |
| 6 | MOBKL1A - Q7L9L4 |
| 7 | CAB39 - Q9Y376 |
Regulation
Activation:
Activated by binding of S100B which releases autoinhibitory N-lobe interactions, enabling ATP to bind and the autophosphorylation of Ser-281. Phosphorylation at Ser-281 increases phosphotransferase activity and interaction with MOBP. Thr-444 then undergoes calcium-dependent phosphorylation by an upstream kinase. Interactions between phosphorylated Thr-444 and the N-lobe promote additional structural changes that complete the activation of the kinase. Phosphorylation at Thr-74 induces interaction with S100B. Thr-444 then undergoes calcium-dependent phosphorylation by an upstream kinase. Interactions between phosphorylated Thr-444 and the N-lobe promote additional structural changes that complete the activation of the kinase. Phosphorylation at Thr-74 induces interaction with S100B.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| AAK1 | Q2M2I8 | S637 | AGHRRILSDVTHSAV | ? |
| NDR1 (NDR; STK38) | Q15208 | S281 | NRRQLAFSTVGTPDY | + |
| NDR1 (NDR; STK38) | Q15208 | T444 | ?Sequence | + |
| p21/Cip1 | P38936 | S146 | GRKRRQTSMTDFYHS | ? |
| PI4KB (suspected) | Q9UBF8 | S277 | RTHQRSKSDATASIS | |
| Rab11flp5 (suspected) | Q9BXF6 | S307 | FTHKRTYSDEANQMR | |
| Rab3IP (Rabin8) | Q96QF0 | S288 | KGHTRNKSTSSAMSG | ? |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
_Specificity_Predicted.gif)
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| K-252a; Nocardiopsis sp. | IC50 > 50 nM | 3813 | 281948 | 22037377 |
| Staurosporine | IC50 > 50 nM | 5279 | 22037377 | |
| Lestaurtinib | Kd = 220 nM | 126565 | 22037378 | |
| KW2449 | Kd = 390 nM | 11427553 | 1908397 | 22037378 |
| Sunitinib | Kd = 410 nM | 5329102 | 535 | 19654408 |
| NVP-TAE684 | Kd = 440 nM | 16038120 | 509032 | 22037378 |
| PDK1/Akt/Flt Dual Pathway Inhibitor | IC50 = 500 nM | 5113385 | 599894 | 22037377 |
| PKR Inhibitor | IC50 = 500 nM | 6490494 | 235641 | 22037377 |
| SB218078 | IC50 = 500 nM | 447446 | 289422 | 22037377 |
| Syk Inhibitor | IC50 = 500 nM | 6419747 | 104279 | 22037377 |
| Dovitinib | Kd = 660 nM | 57336746 | 22037378 | |
| SU14813 | Kd = 700 nM | 10138259 | 1721885 | 22037378 |
| JAK3 Inhibitor VI | IC50 > 1 µM | 16760524 | 22037377 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 |
Disease Linkage
General Disease Association:
Cancer, Infectious disease
Specific Diseases (Non-cancerous):
HIV
Comments:
Like NDR2, NDR1 is found incorporated into HIV-1 particles, and it is speculated that they may play a role in the HIV-1 life cycle.
Specific Cancer Types:
B-Cell lymphomas
Comments:
NDR1 is an important regulator of Myc's oncogenic activity in human B-cell lymphomas. It also regulates G/S cell cycle transition.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -62, p<0.006); and Brain oligodendrogliomas (%CFC= -52, p<0.02). The COSMIC website notes an up-regulated expression score for NDR1 in diverse human cancers of 539, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 64 for this protein kinase in human cancers was 1.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. T74A, K118A, S281A, and T444A mutations in NDR1 are involved in decreased or loss of autophosphorylation and phosphotransferase activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25371 diverse cancer specimens. This rate is very similar (+ 5% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.47 % in 1093 large intestine cancers tested; 0.39 % in 602 endometrium cancers tested; 0.32 % in 805 skin cancers tested; 0.12 % in 1941 lung cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,654 cancer specimens
Comments:
Only 6 deletions, and no insertions or complex mutations are noted on the COSMIC website.

