Nomenclature
Short Name:
NEK2
Full Name:
Serine-threonine-protein kinase Nek2
Alias:
- EC 2.7.11.1
- HSPK 21
- NimA-related protein kinase 2
- NLK1
- Kinase Nek2
- NEK2A
- NIMA (never in mitosis gene a)-related kinase 2
- NimA-like protein kinase 1
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
NEK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
51,763
# Amino Acids:
445
# mRNA Isoforms:
4
mRNA Isoforms:
51,763 Da (445 AA; P51955); 50,909 Da (437 AA; P51955-4); 44,906 Da (384 AA; P51955-2); 37,956 Da (326 AA; P51955-3)
4D Structure:
Isoform 1 and isoform 2 form homo- and heterodimers. Interacts with TERF1, SGOL1 and MAD1L1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
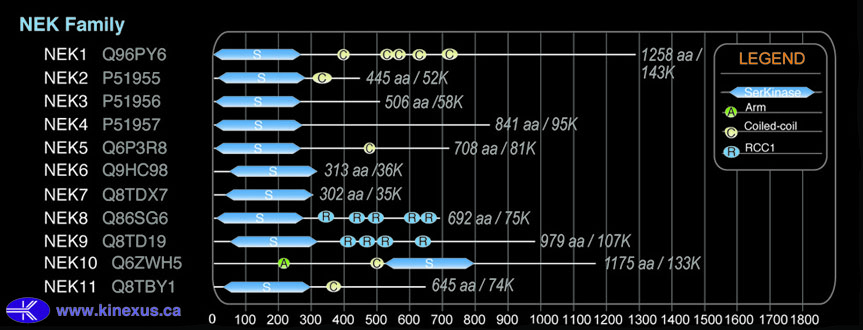
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 8 | 271 | Pkinase |
| 303 | 356 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K437.
Serine phosphorylated:
S171+, S184-, S241-, S296, S300, S304, S356, S365, S378, S397, S400, S402, S406, S428, S438.
Threonine phosphorylated:
T170+, T175+, T179-.
Ubiquitinated:
K143, K152, K156, K362, K391, K407.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 19
19
1038
29
1668
 0.2
0.2
11
15
12
 0.8
0.8
45
5
47
 29
29
1617
95
3868
 8
8
439
25
399
 33
33
1791
84
7402
 3
3
159
31
333
 100
100
5510
38
8624
 7
7
381
17
315
 0.3
0.3
18
80
22
 0.2
0.2
13
27
15
 8
8
430
159
435
 0.3
0.3
15
27
14
 0.3
0.3
17
12
16
 0.3
0.3
14
14
16
 0.1
0.1
7
14
9
 6
6
331
117
3291
 0.4
0.4
20
17
26
 0.3
0.3
16
84
15
 6
6
321
109
358
 0.3
0.3
17
20
24
 0.2
0.2
12
19
11
 0.3
0.3
19
19
20
 7
7
369
18
244
 1
1
53
19
51
 92
92
5053
56
9368
 0.8
0.8
44
31
27
 0.4
0.4
22
18
23
 0.3
0.3
17
18
22
 0.8
0.8
44
28
36
 4
4
234
24
228
 11
11
585
36
939
 15
15
837
72
1197
 11
11
603
57
572
 18
18
977
35
1699
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 85.1
85.1
85.1
100 98.4
98.4
99.3
- -
-
-
93 -
-
-
99 62.6
62.6
65.4
94 -
-
-
- 87.6
87.6
94.2
88.5 89.4
89.4
95.1
88 -
-
-
- 69.7
69.7
81
- 77.1
77.1
88.8
77 73.9
73.9
86.7
73 25.8
25.8
39
69 -
-
-
- 30.2
30.2
44.4
36 -
-
-
- -
-
-
- 54.3
54.3
72.2
- -
-
-
- -
-
-
- -
-
-
- 27.2
27.2
44.9
- 22.7
22.7
43.8
- 24.4
24.4
36.1
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | NDC80 - O14777 |
| 2 | CDC27 - P30260 |
| 3 | MAPK1 - P28482 |
| 4 | PPP1CA - P62136 |
| 5 | MAD1L1 - Q9Y6D9 |
| 6 | MAD2L1 - Q13257 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
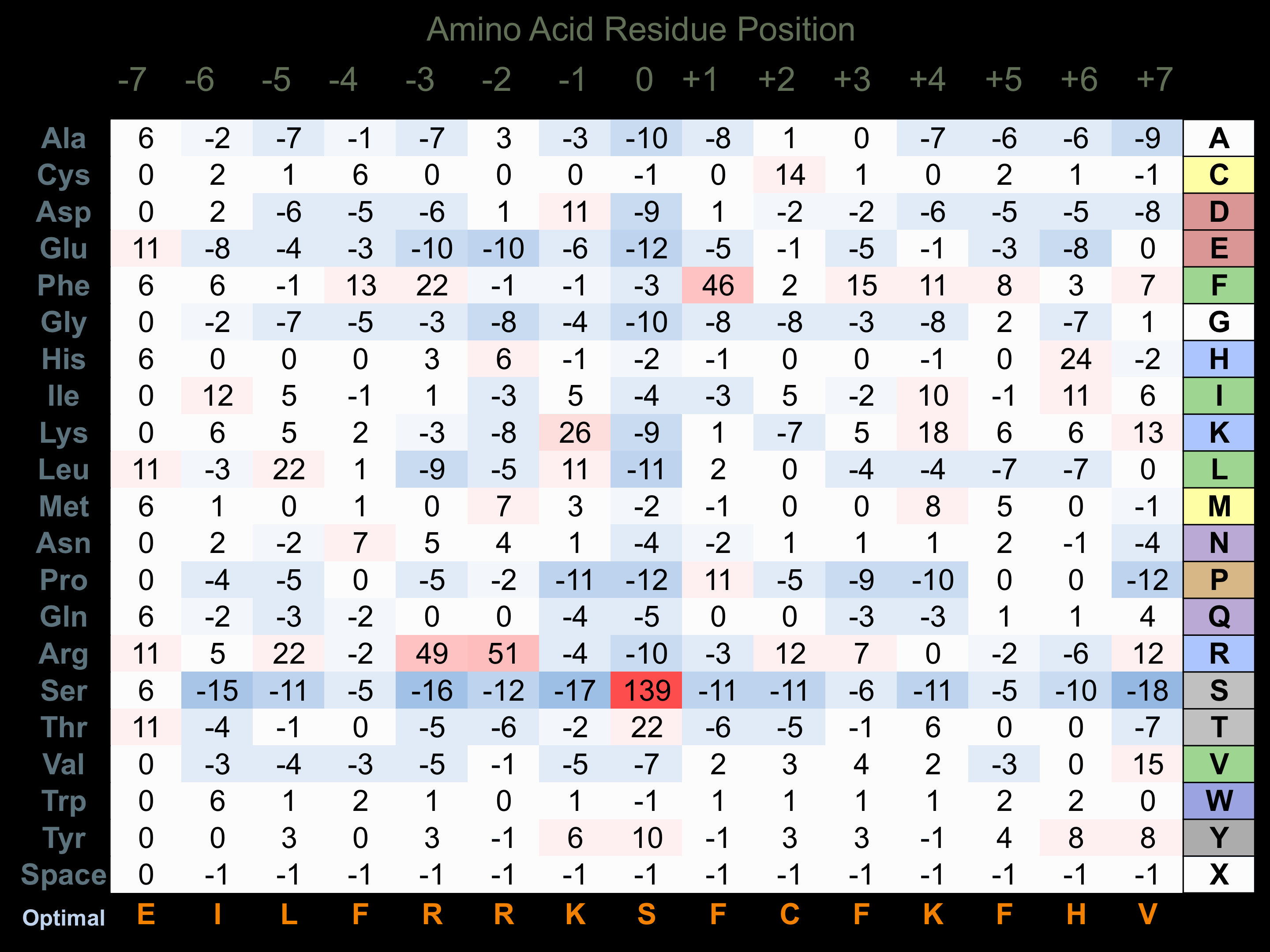
Matrix Type:
Experimentally derived from alignment of 14 known protein substrate phosphosites and 21 peptides phosphorylated by recombinant NEK2 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, physiological development, and retinal disorders
Specific Diseases (Non-cancerous):
Van Der Woude syndrome (LDS); Retinitis pigmentosa 67 (RP67); Leber congenital amaurosis (CRB)
Comments:
Van Der Woude Syndrome (LDS) is a development disease affecting the structure of the face, often characterized by cleft lip and, or a cleft palate. LDS can affec the testis, brain, and salivary glands. Retinitis Pigmentosa 67 (RP67) is normally expressed in the cells at the back of the eye. RP67 is typically characterized by loss of rod photoreceptor cells followed by loss of cone cells, leading to night vision blindness, and loss of the midperipheral vision field. Leber Congenital Amaurosis (CRB) is a rare condition characterized by vision impairment during early infancy, photophobia, nystagmus (uncontrolled eye movement), far-sightedness, night blindness, abnormal pupil reactivity, and abnormal scaring. CRB affects the retina, but can also affect the eye and testis tissues.
Specific Cancer Types:
Plexiform Neurofibromas; Polyploidy; Breast cancer
Comments:
NEK2 is linked to Plexiform Neurofibromas, which are related to the disorders neurofibroma and neurofibromatosis disorders, and can affect skin, salivary gland, and bone tissues. Polyploidy is the abnormal duplication of chromosomes above the typical diploid level. Polyploidy is linked to breast cancer and melanoma, and most often affects smooth muscle, liver, and breast tissues. NEK2 has been associated with both primary and secondary breast tumours.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +162, p<0.013); Breast epithelial carcinomas (%CFC= +309, p<0.068); Breast non-basal-like cancer (BLC) (%CFC= +76, p<0.0001); Breast sporadic basal-like cancer (BLC) (%CFC= +102, p<0.0001); Cervical cancer (%CFC= -79, p<0.0001); Colon mucosal cell adenomas (%CFC= +147, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= +227, p<0.006); Ovary adenocarcinomas (%CFC= +428, p<0.0009); Prostate cancer (%CFC= -58, p<0.025); and Vulvar intraepithelial neoplasia (%CFC= +307, p<0.0004). The COSMIC website notes an up-regulated expression score for NEK2 in diverse human cancers of 878, which is 1.9-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 11 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. The ability for NEK2 to activate NEK11 is lost with a K37R mutation. The autophosphorylation of NEK2 can be inhibited with a D141A mutation. NEK2 kinase phosphotransferase activity can be increased 2-fold with a T170E, or S171D or T175E mutation, while the phosphotransferase activity can be decreased two-fold with a T175A mutation. Complete kinase phosphotransferase activity can be lost with a T179A, T179E, S241A or S241D mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 25158 diverse cancer specimens. This rate is only -38 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.29 % in 864 skin cancers tested; 0.22 % in 603 endometrium cancers tested; 0.1 % in 1512 liver cancers tested; 0.09 % in 238 bone cancers tested; 0.09 % in 1270 large intestine cancers tested; 0.08 % in 273 cervix cancers tested; 0.08 % in 1490 breast cancers tested; 0.05 % in 1768 lung cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: S296L (4).
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

