Nomenclature
Short Name:
CDC7
Full Name:
Cell division cycle 7-related protein kinase
Alias:
- CDC7- related kinase
- CDC7L1
- HuCdc7
- Kinase CDC7
- MuCdc7
- Cell division cycle 7
- EC 2.7.11.1
- HsCdc7
- Hsk1
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
CDC7
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
63,888
# Amino Acids:
574
# mRNA Isoforms:
1
mRNA Isoforms:
63,888 Da (574 AA; O00311)
4D Structure:
Forms a complex with either DBF4/DBF4A or DBF4B, leading to the activation of the kinase activity.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
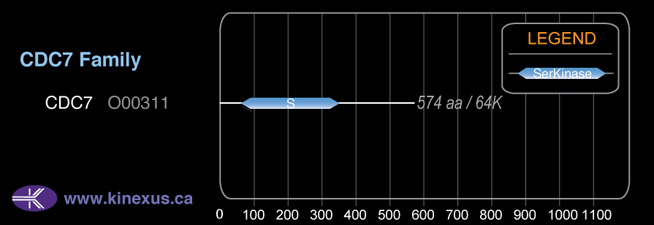
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 58 | 569 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K331.
Serine phosphorylated:
S16, S27, S181, S277, S297, S302, S484, S490, S498, S509, S512, S556, S573.
Threonine phosphorylated:
T376-, T386, T472, T503, T559.
Tyrosine phosphorylated:
Y415.
Ubiquitinated:
K30, K212.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 57
57
990
16
1335
 1.3
1.3
22
10
17
 2
2
36
11
38
 38
38
658
57
1394
 24
24
414
14
404
 15
15
255
45
402
 16
16
269
19
450
 100
100
1727
32
4434
 14
14
243
10
214
 2
2
40
45
26
 2
2
29
25
32
 28
28
488
105
591
 7
7
113
23
116
 2
2
43
9
29
 2
2
33
22
38
 2
2
38
8
37
 2
2
26
108
17
 2
2
33
19
35
 2
2
33
54
22
 25
25
424
56
450
 3
3
60
21
48
 9
9
151
23
160
 3
3
57
21
40
 13
13
223
19
273
 6
6
108
21
127
 96
96
1663
38
4104
 4
4
72
26
74
 2
2
38
19
33
 2
2
43
19
49
 3
3
50
14
45
 57
57
989
18
660
 24
24
416
21
489
 37
37
636
51
1012
 35
35
611
31
624
 4
4
65
22
57
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 0
0
0
98 96.8
96.8
97.7
97 -
-
-
89 -
-
-
97 68.9
68.9
71.5
91 -
-
-
- 78
78
84.8
81 -
-
-
81.5 -
-
-
- 61.3
61.3
65.6
- 67.2
67.2
78.7
70 55.4
55.4
66.9
70 -
-
-
64 -
-
-
- -
-
-
34 -
-
-
- -
-
-
- 36.7
36.7
54.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 22.1
22.1
39.2
26 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | ORC5L - O43913 |
| 2 | MCM4 - P33991 |
| 3 | MCM2 - P49736 |
| 4 | CDK4 - P11802 |
| 5 | DBF4 - Q9UBU7 |
| 6 | MCM5 - P33992 |
| 7 | ORC6L - Q9Y5N6 |
| 8 | MCM3 - P25205 |
| 9 | MCM7 - P33993 |
| 10 | SUMO1 - P63165 |
| 11 | DBF4B - Q8NFT6 |
| 12 | CDK2 - P24941 |
| 13 | CHAF1A - Q13111 |
| 14 | MCM6 - Q14566 |
Regulation
Activation:
Activated by phosphorylation at Thr-376 in the kinase activation loop by CDK1 and CDK2. It also requires binding to activator of S-phase kinase (ASK)/DBF4B to be fully stimulated.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| MCM2 | P49736 | S108 | DVEELTASQREAAER | + |
| MCM2 | P49736 | S139 | RRGLLYDSDEEDEER | + |
| MCM2 | P49736 | S220 | NVFKERISDMCKENR | |
| MCM2 | P49736 | S27 | GNDPLTSSPGRSSRR | ? |
| MCM2 | P49736 | S31 | LTSSPGRSSRRTDAL | |
| MCM2 | P49736 | S4 | ____MAESSESFTMA | |
| MCM2 | P49736 | S40 | RRTDALTSSPGRDLP | ? |
| MCM2 | P49736 | S41 | RTDALTSSPGRDLPP | + |
| MCM2 | P49736 | S53 | LPPFEDESEGLLGTE | ? |
| MCM2 | P49736 | S7 | _MAESSESFTMASSP | |
| MCM2 | P49736 | T59 | ESEGLLGTEGPLEEE |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
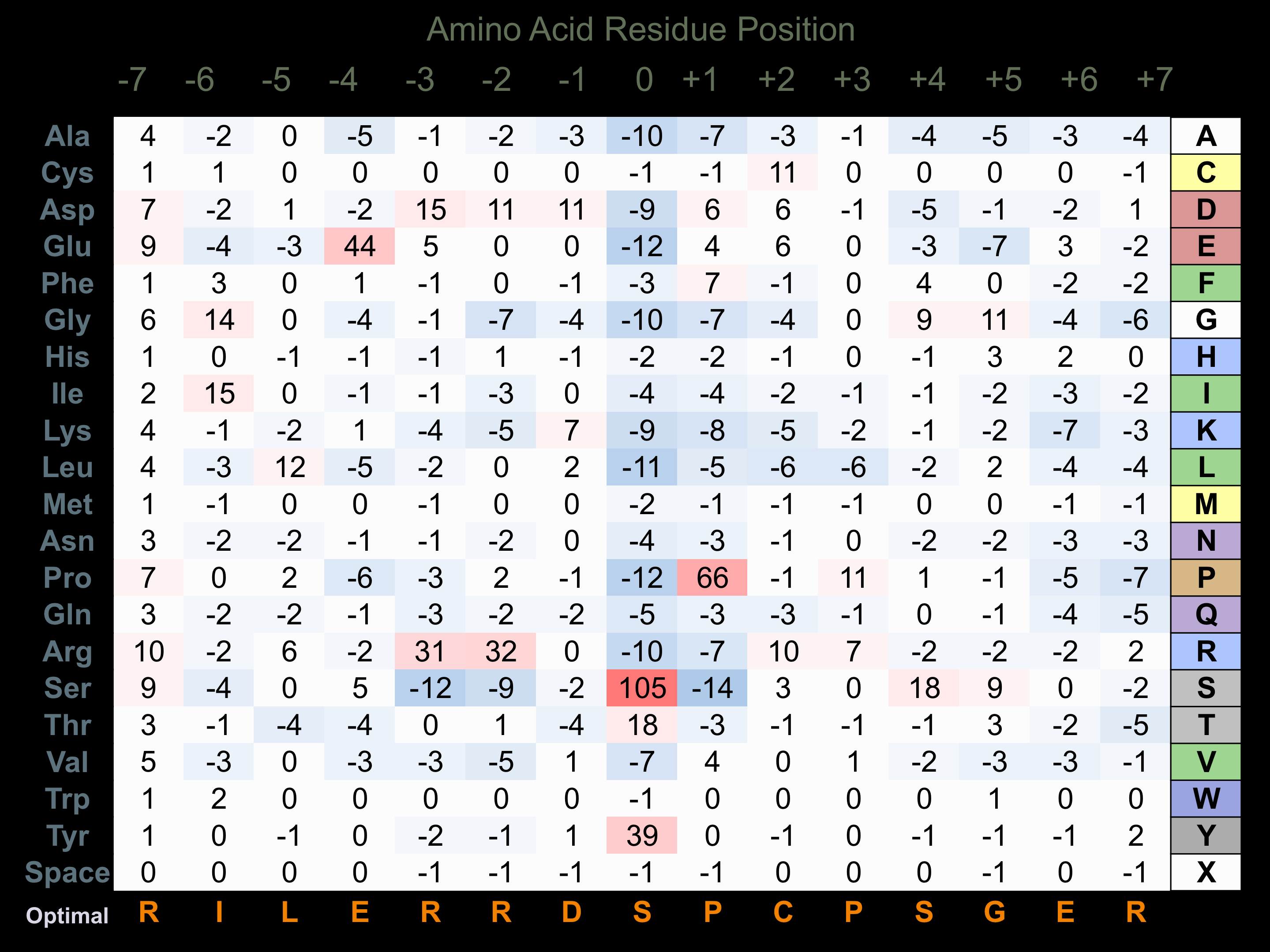
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| PHA-767491 | IC50 = 10 nM | 11715767 | 225519 | 18201066 |
| CHEMBL1258913 | IC50 = 1.97 µM | 11847343 | 1258913 | 20817473 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Breast; Pancreatic Cancer
Comments:
CDC7 may be an oncoprotein (OP). over-expression of CDC7 may be associated with neoplastic tumour transformation. RNAi inhibition of CDC7 induces an abortive S phase and apoptotic cell death in p53-mutant ErbB2-overexpressing and triple-negative (ER-/PR-/ErbB2-) breast cancer cell lines, but not in untransformed breast epithelial cells. Consequently, CDC7 appears to represent a potent and highly specific anticancer target in ErbB2-overexpressing and triple-negative breast cancers. It is also a suitable target for treatment of other types of cancer, including pancreatic cancer.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +53, p<0.066); Breast non-basal-like cancer (BLC) (%CFC= +53, p<0.0001); Breast sporadic basal-like cancer (BLC) (%CFC= +77, p<0.0001); Cervical cancer (%CFC= -64, p<0.0001); Classical Hodgkin lymphomas (%CFC= +66, p<0.003); Clear cell renal cell carcinomas (cRCC) (%CFC= +52, p<0.043); Colon mucosal cell adenomas (%CFC= +80, p<0.0001); Large B-cell lymphomas (%CFC= +110, p<0.0002); Oral squamous cell carcinomas (OSCC) (%CFC= +49, p<0.004); Ovary adenocarcinomas (%CFC= +240, p<0.0008); andVulvar intraepithelial neoplasia (%CFC= +216, p<0.0001). The COSMIC website notes an up-regulated expression score for CDC7 in diverse human cancers of 428, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 7 for this protein kinase in human cancers was 0.1-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25496 diverse cancer specimens. This rate is only 16 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.67 % in 1270 large intestine cancers tested; 0.36 % in 629 stomach cancers tested; 0.32 % in 603 endometrium cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: L28* (8), N31K (2).
Comments:
Twenty five N31fs*51 frameshift deletions, 3 N31fs*6 insertions noted on the COSMIC website. These mutations results in a truncated protein that is missing the kinase catalytic domain.

