Nomenclature
Short Name:
p38d
Full Name:
Mitogen-activated protein kinase 13
Alias:
- EC 2.7.1.37
- EC 2.7.11.24
- p38 MAPK-delta
- P38delta
- PRKM13
- Stress-activated protein kinase-4; SAPK4; p38d MAPK
- MAP kinase p38 delta
- MAPK13
- Mitogen-activated protein kinase p38 delta
- MK13
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
MAPK
SubFamily:
p38
Specific Links
Structure
Mol. Mass (Da):
42,090
# Amino Acids:
365
# mRNA Isoforms:
2
mRNA Isoforms:
42,090 Da (365 AA; O15264); 28,779 Da (257 AA; O15264-2)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
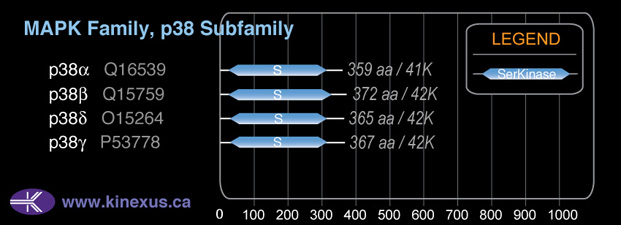
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 25 | 308 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S47, S350, S357, S361.
Threonine phosphorylated:
T180+, T185.
Tyrosine phosphorylated:
Y10, Y182+.
Ubiquitinated:
K226.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 49
49
993
29
1238
 9
9
185
15
91
 4
4
82
13
63
 39
39
800
93
2625
 42
42
863
25
687
 8
8
155
78
124
 20
20
402
31
404
 84
84
1711
44
3070
 45
45
919
17
642
 4
4
76
81
51
 3
3
57
30
58
 36
36
735
164
672
 3
3
59
35
38
 2
2
37
9
33
 6
6
113
26
74
 10
10
205
15
81
 2
2
44
120
37
 10
10
207
22
144
 3
3
55
98
47
 38
38
768
109
706
 2
2
37
22
44
 3
3
57
27
43
 7
7
138
15
144
 10
10
201
22
116
 4
4
79
24
65
 82
82
1664
62
3167
 13
13
272
38
958
 5
5
108
22
77
 3
3
59
20
65
 6
6
121
28
92
 42
42
849
24
778
 100
100
2032
36
3982
 2
2
38
70
149
 54
54
1098
52
796
 89
89
1812
35
1566
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 97.3
97.3
97.5
100 64.8
64.8
78.5
92 -
-
-
95 -
-
-
95 94
94
97.3
94 -
-
-
- 92.9
92.9
97.3
93 91.8
91.8
96.2
93 -
-
-
- 84.8
84.8
91.4
- 44.8
44.8
63.6
79.5 59.7
59.7
75.9
76 60
60
77
67 -
-
-
- 56.4
56.4
73.2
- -
-
-
- 51.2
51.2
72.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by phosphorylation at Tyr-182 and probably Thr-180 by MAP2K3 (MKK3) and MAP2K6 (MKK6).
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Cyclin D3 (CCND3) | P30281 | T283 | QGPSQTSTPTDVTAI | |
| DLG1 (SAP97) | Q12959 | S122 | TPPQEHISPQITNEV | |
| DLG1 (SAP97) | Q12959 | S158 | FVSHSHISPIKPTEA | |
| DLG1 (SAP97) | Q12959 | T209 | VNTDSLETPTYVNGT | |
| DLG4 (PSD-95) | P78352 | T287 | TDYPTAMTPTSPRRY | |
| eEF2K | O00418 | S359 | GTEEKCGSPRVRTLS | - |
| eEF2K | O00418 | S396 | TFDSLPSSPSSATPH | |
| MAFA | Q8NHW3 | S335 | AGFPREPSPPQAGPG | |
| MAFA | Q8NHW3 | T113 | NPEALNLTPEDAVEA | |
| MAFA | Q8NHW3 | T57 | LSSTPLSTPCSSVPS | |
| PKD1 (PRKCM) | Q15139 | S205 | GVRRRRLSNVSLTGV | - |
| PKD1 (PRKCM) | Q15139 | S208 | RRRLSNVSLTGVSTI | - |
| PKD1 (PRKCM) | Q15139 | S249 | GREKRSNSQSYIGRP | + |
| PKD1 (PRKCM) | Q15139 | S397 | EDANRTISPSTSNNI | - |
| PKD1 (PRKCM) | Q15139 | S401 | RTISPSTSNNIPLMR | |
| PKD1 (PRKCM) | Q15139 | S742 | GEKSFRRSVVGTPAY | + |
| STMN1 | P16949 | S25 | QAFELILSPRSKESV | |
| STMN1 | P16949 | S38 | SVPEFPLSPPKKKDL | |
| Tau iso8 | P10636-8 | S396 | GAEIVYKSPVVSGDT | |
| Tau iso8 | P10636-8 | S404 | PVVSGDTSPRHLSNV | |
| Tau iso8 | P10636-8 | T212 | TPGSRSRTPSLPTPP | |
| Tau iso8 | P10636-8 | T217 | SRTPSLPTPPTREPK |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
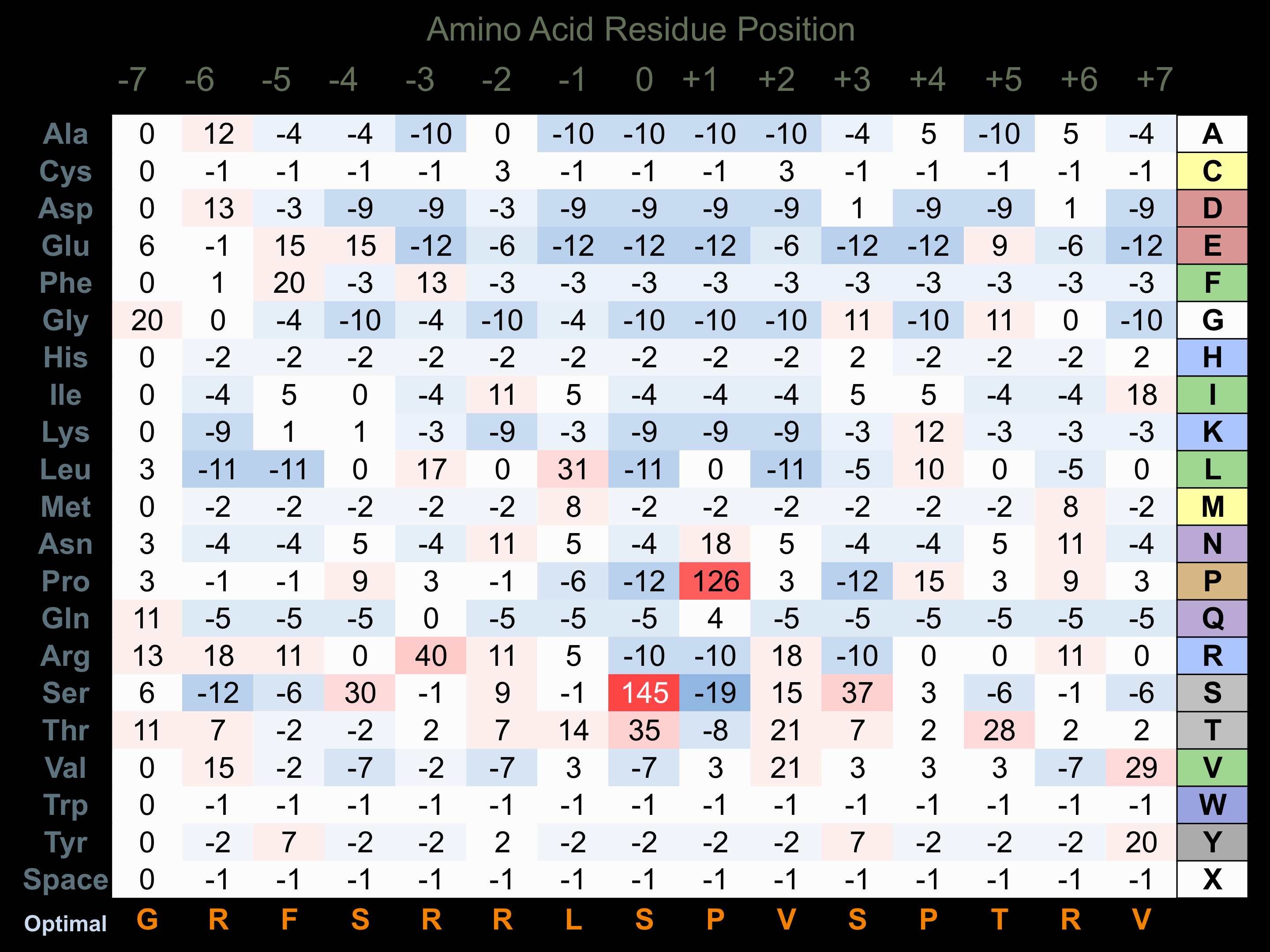
Matrix Type:
Experimentally derived from alignment of 28 known protein substrate phosphosites. Note that additional binding sites on p38 substrates with D motifs (consensus= K/R-k-x-s-l/p-l-l-l-p-p or p-x-L/v/i-x-p-p-x-x-x-x-l-l-x-r/k-k/r-R/k-K/r) facilitate higher selectivity for phosphorylation by this protein kinase.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, endocrine, and neurological disorders
Specific Diseases (Non-cancerous):
Diabetes mellitus; Alzheimer's disease (AD)
Specific Cancer Types:
Cholangiocarcinomas (CCA, CC)
Comments:
The rare liver cancer Cholangiocarcinoma (CCA, CC) arises from transformed epithelial cells and may include symptoms such as abnormally dark urine, and itching alongside tumour development. CC has been noted to have a significant up-regulation of p38d compared to normal bile duct tissue
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -45, p<0.009); Bladder carcinomas (%CFC= +116, p<0.0001); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= +67, p<0.084); Cervical cancer (%CFC= +75, p<0.002); Cervical cancer stage 1B (%CFC= -60, p<0.021); Cervical cancer stage 2B (%CFC= -83, p<0.015); Colorectal adenocarcinomas (early onset) (%CFC= -48, p<0.0002); Lung adenocarcinomas (%CFC= +92, p<0.0001); Oral squamous cell carcinomas (OSCC) (%CFC= -49, p<0.0009); Ovary adenocarcinomas (%CFC= +89, p<0.002); Papillary thyroid carcinomas (PTC) (%CFC= +73, p<0.0006); Skin melanomas - malignant (%CFC= -64, p<0.0002); Skin squamous cell carcinomas (%CFC= +67, p<0.029); and Vulvar intraepithelial neoplasia (%CFC= +68, p<0.004). The COSMIC website notes an up-regulated expression score for p38d in diverse human cancers of 538, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 37 for this protein kinase in human cancers was 0.6-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. The phosphotransferase activity of p38d can be abrogated with a T180A or a Y182A mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25578 diverse cancer specimens. This rate is only 20 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.85 % in 805 skin cancers tested; 0.47 % in 1119 large intestine cancers tested.
Frequency of Mutated Sites:
None > 7 in 20,862 cancer specimens
Comments:
Only 1 deletion, and no insertions or complex mutations are noted on the COSMIC website.

