Nomenclature
Short Name:
LOK
Full Name:
Serine-threonine-protein kinase 10
Alias:
- EC 2.7.11.1
- GEK2
- STK10
- Lymphocyte-oriented kinase
- PRO2729
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
SLK
Specific Links
Structure
Mol. Mass (Da):
112,135
# Amino Acids:
968
# mRNA Isoforms:
1
mRNA Isoforms:
112,135 Da (968 AA; O94804)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
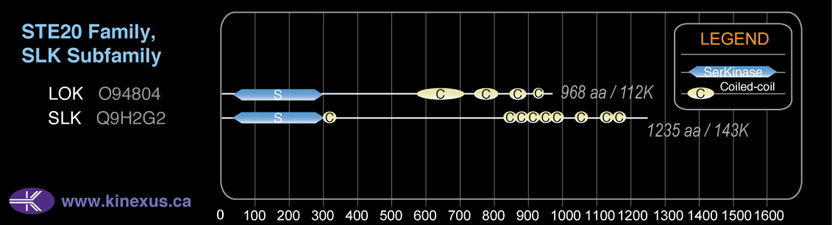
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 36 | 294 | Pkinase |
| 573 | 704 | Coiled-coil |
| 738 | 803 | Coiled-coil |
| 844 | 887 | Coiled-coil |
| 920 | 940 | Coiled-coil |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K690.
Serine phosphorylated:
S13, S20, S179, S191, S392, S417, S438, S444, S448, S450, S454, S455, S485, S495, S514, S545, S549, S679, S844, S951, S954, S968.
Threonine phosphorylated:
T14, T166, T185, T195-, T208, T391, T459, T530, T688, T952.
Tyrosine phosphorylated:
Y197, Y962.
Ubiquitinated:
K56, K65, K151, K159, K181, K184, K621.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 54
54
1308
35
1302
 2
2
47
13
28
 26
26
641
16
588
 16
16
394
120
877
 32
32
766
28
479
 2
2
44
83
23
 8
8
192
38
427
 52
52
1253
45
3336
 21
21
498
14
318
 4
4
88
110
172
 12
12
288
34
365
 36
36
861
172
665
 45
45
1093
38
1394
 2
2
45
9
42
 10
10
245
28
303
 2
2
45
20
27
 4
4
101
201
178
 27
27
655
22
1589
 5
5
121
106
659
 23
23
566
134
561
 14
14
344
26
427
 44
44
1079
30
1044
 26
26
639
18
645
 19
19
455
22
400
 29
29
706
26
662
 53
53
1286
75
2605
 23
23
551
38
688
 16
16
391
22
541
 15
15
369
22
523
 3
3
81
42
57
 47
47
1145
24
589
 100
100
2425
41
4934
 16
16
390
84
437
 45
45
1094
78
794
 7
7
167
48
190
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.1
98.1
98.7
99 24
24
35.9
98 -
-
-
90 -
-
-
97 91.5
91.5
95.5
92 -
-
-
- 88.4
88.4
94.2
89 44.9
44.9
61.1
86 -
-
-
- 52
52
62.9
- 24.1
24.1
35.3
74 24.1
24.1
36.3
69 24
24
36
64 -
-
-
- 24.7
24.7
41.1
50 -
-
-
- 22.7
22.7
34.2
40 -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
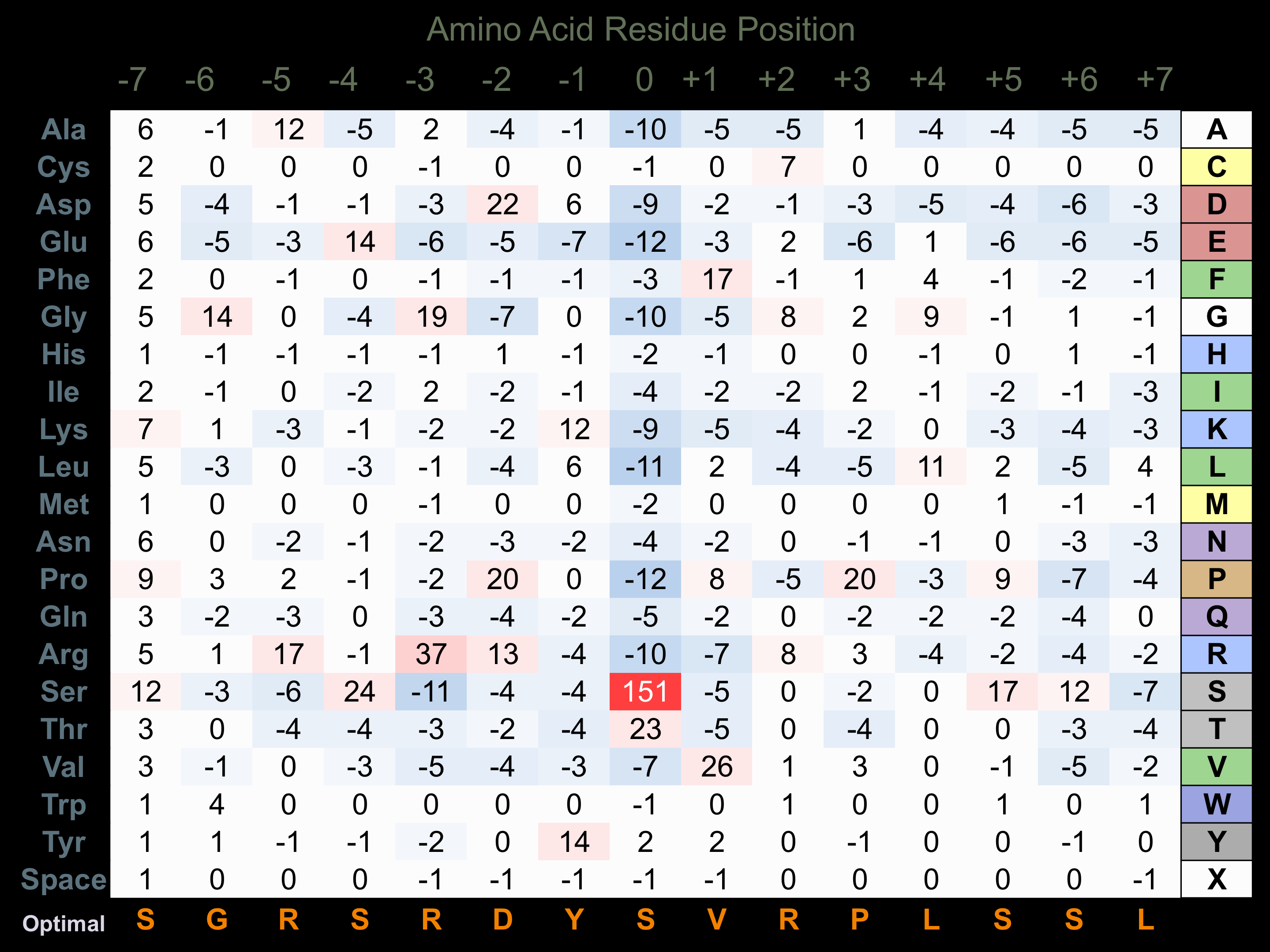
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer, gastrointestinal disorders
Specific Diseases (Non-cancerous):
Cholecystolithiasis
Comments:
Cholecystolithiasis is a gastrointestinal disease with a common name, gallstone, which developes from bile accumulation in the gallbladder.
Specific Cancer Types:
Germ cell tumours (GCT); Testicular germ cell tumours (TGCT); Male germ cell tumours, somatic; Testicular tumours, somatic; Spermatocytic seminomas, somatic
Comments:
Germ Cell tumours (GCT) are rare diseases that arise from germ cells, and that can affect the testis, ovaries, and lymph node tissues. Testicular germ cell tumour (TGCT) which has a relation to teratoma and testicular cancer disorders can affect the testis, and lymph nodes. A K65I mutation in LOK with TGCT can induce loss of kinase phosphotransferase activity. Male Germ Cell Tumour, Somatic is a rare condition which has a relation to testicular cancer and testicular germ cell tumour. Spermatocytic Seminoma, Somatic is a reproductive disease with a relation to polyhydramnios and papilloma (which forms a growth on epithelial tissue).
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= +98, p<0.002); Brain glioblastomas (%CFC= -89, p<0.042); Brain oligodendrogliomas (%CFC= -71, p<0.09); Clear cell renal cell carcinomas (cRCC) (%CFC= +88, p<0.016); Large B-cell lymphomas (%CFC= +51, p<0.032); Oral squamous cell carcinomas (OSCC) (%CFC= +128, p<0.002); Skin fibrosarcomas (%CFC= +59, p<0.063); and Skin melanomas - malignant (%CFC= +117, p<0.0002). The COSMIC website notes an up-regulated expression score for LOK in diverse human cancers of 258, which is 0.6-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 10 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 25412 diverse cancer specimens. This rate is only -6 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 1.03 % in 10 peritoneum cancers tested; 0.38 % in 1229 large intestine cancers tested; 0.26 % in 864 skin cancers tested; 0.18 % in 589 stomach cancers tested; 0.15 % in 603 endometrium cancers tested; 0.13 % in 548 urinary tract cancers tested; 0.12 % in 1608 lung cancers tested; 0.11 % in 273 cervix cancers tested; 0.09 % in 710 oesophagus cancers tested; 0.08 % in 891 ovary cancers tested; 0.06 % in 1512 liver cancers tested; 0.06 % in 1490 breast cancers tested; 0.04 % in 959 upper aerodigestive tract cancers tested; 0.04 % in 238 bone cancers tested.
Frequency of Mutated Sites:
None > 4 in 20,694 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

