Nomenclature
Short Name:
PFTAIRE1
Full Name:
PFTAIRE-1
Alias:
- CDK14
- Serine/threonine protein kinase PFTAIRE-1
- EC 2.7.11.22
- KIAA0834
- PFT1
- PFTAIRE protein kinase 1
- PFTK1
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
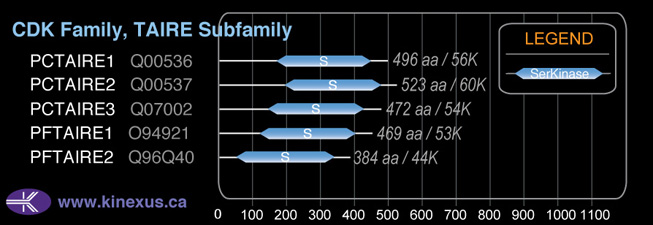
CDK
SubFamily:
TAIRE
Structure
Mol. Mass (Da):
53057
# Amino Acids:
469
# mRNA Isoforms:
3
mRNA Isoforms:
53,057 Da (469 AA; O94921); 50,662 Da (451 AA; O94921-2); 47,760 Da (423 AA; O94921-3)
4D Structure:
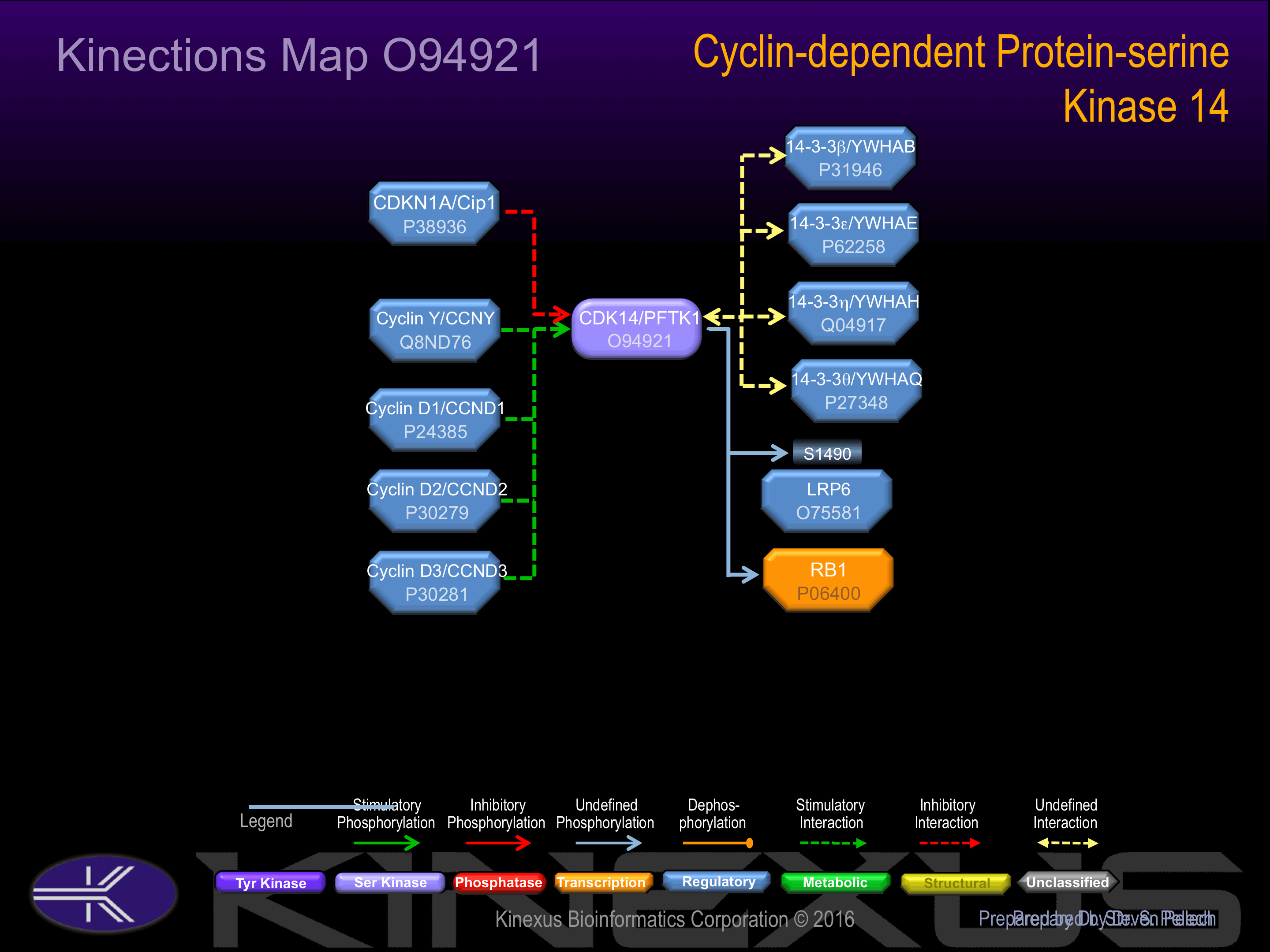
Interacts with CCNY; CCNY mediates its recruitment to the plasma membrane and promotes phosphorylation of LRP6. Interacts with CCDN3 and CDKN1A. Interacts with SEPT8. Interacts with 14-3-3 proteina YWHAB, YWHAE, YWHAH and YWHAQ.
1D Structure:
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 135 | 419 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S24, S26, S78, S95, S106, S112, S119, S120, S122, S123, S126, S134, S145.
Threonine phosphorylated:
T76, T105, T125, T202.
Tyrosine phosphorylated:
Y135, Y146.
Ubiquitinated:
K271.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1221
44
1158
 3
3
39
17
29
 5
5
67
11
53
 34
34
417
143
515
 50
50
612
44
559
 4
4
47
98
78
 10
10
121
51
336
 32
32
385
46
508
 32
32
395
17
334
 9
9
112
136
112
 7
7
84
36
77
 56
56
678
191
639
 8
8
95
34
60
 7
7
88
12
77
 13
13
156
21
148
 5
5
55
25
84
 8
8
97
133
61
 7
7
80
20
60
 5
5
65
122
45
 37
37
454
165
461
 12
12
144
29
103
 9
9
105
32
73
 11
11
133
13
63
 9
9
110
20
74
 5
5
65
28
69
 52
52
640
88
545
 6
6
74
36
55
 5
5
67
21
49
 7
7
85
20
57
 11
11
134
56
109
 23
23
278
24
275
 40
40
484
46
545
 5
5
61
95
211
 73
73
896
104
696
 40
40
493
61
589
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.3
99.3
99.5
99 99.7
99.7
99.7
100 -
-
-
98 -
-
-
100 99.1
99.1
100
99 -
-
-
- 98.5
98.5
99.3
98.5 43
43
59
98 -
-
-
- 60.7
60.7
63.3
- 58.5
58.5
59.5
96 85.2
85.2
89.3
94 51.5
51.5
63.1
95 -
-
-
- 48.4
48.4
61.2
68 50
50
63.3
- -
-
-
57 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 30.7
30.7
42.8
- 28.5
28.5
43.9
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Kinase catalytic activity stimulated by associated cyclins CCDN3 and CCNY.
Inhibition:
Repressed by CDKN1A.
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
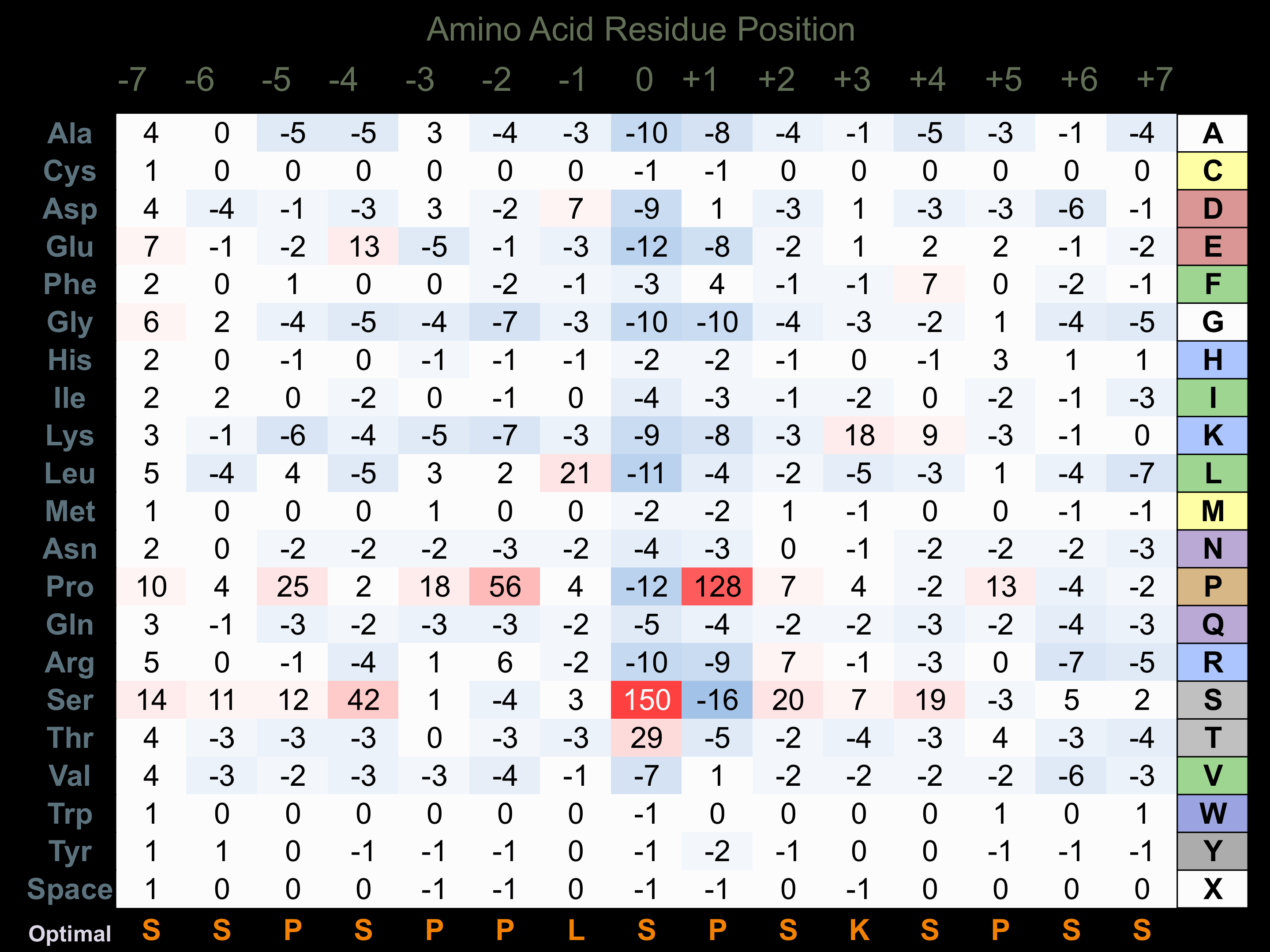
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| R547 | Kd = 11 nM | 6918852 | 22037378 | |
| AT7519 | Kd = 16 nM | 11338033 | 22037378 | |
| AST-487 | Kd = 17 nM | 11409972 | 574738 | 18183025 |
| Foretinib | Kd = 31 nM | 42642645 | 1230609 | 22037378 |
| Alvocidib | Kd = 110 nM | 9910986 | 428690 | 18183025 |
| Staurosporine | Kd = 160 nM | 5279 | 18183025 | |
| Sunitinib | Kd = 270 nM | 5329102 | 535 | 18183025 |
| Nintedanib | Kd = 470 nM | 9809715 | 502835 | 22037378 |
| AC1NS7CD | Kd = 690 nM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 690 nM | 3025986 | 296468 | 18183025 |
| Lestaurtinib | Kd = 720 nM | 126565 | 22037378 | |
| A674563 | Kd = 790 nM | 11314340 | 379218 | 22037378 |
| KW2449 | Kd = 870 nM | 11427553 | 1908397 | 22037378 |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| SU14813 | Kd = 1.1 µM | 10138259 | 1721885 | 22037378 |
| PHA-665752 | Kd = 1.3 µM | 10461815 | 450786 | 22037378 |
| Bosutinib | Kd = 1.4 µM | 5328940 | 288441 | 22037378 |
| Enzastaurin | Kd = 1.5 µM | 176167 | 300138 | 22037378 |
| TG101348 | Kd = 2.6 µM | 16722836 | 1287853 | 22037378 |
| Sorafenib | Kd = 2.9 µM | 216239 | 1336 | 18183025 |
| JNJ-7706621 | Kd = 3.6 µM | 5330790 | 191003 | 18183025 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Oesophageal squamous cell carcinomas
Comments:
Positive expression of PFTAIRE1 when compared to its negative expression predicts resistance to chemotherapy and poorer prognosis in patients with oesophageal squamous cell carcinomas over a 5-year period.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= +359, p<0.002); Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -47, p<0.007); Cervical cancer (%CFC= -46, p<0.004); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= -73, p<0.0004); Head and neck squamous cell carcinomas (HNSCC) (%CFC= +102, p<0.002); Oral squamous cell carcinomas (OSCC) (%CFC= +178, p<0.01); and Skin melanomas - malignant (%CFC= -47, p<0.039). The COSMIC website notes an up-regulated expression score for PFTAIRE1 in diverse human cancers of 538, which is 1.2-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 9 for this protein kinase in human cancers was 0.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.12 % in 24939 diverse cancer specimens. This rate is a modest 1.58-fold higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.57 % in 1270 large intestine cancers tested; 0.49 % in 864 skin cancers tested; 0.4 % in 589 stomach cancers tested; 0.32 % in 603 endometrium cancers tested; 0.24 % in 710 oesophagus cancers tested; 0.22 % in 1822 lung cancers tested; 0.17 % in 127 biliary tract cancers tested; 0.16 % in 548 urinary tract cancers tested; 0.16 % in 273 cervix cancers tested; 0.15 % in 1316 breast cancers tested; 0.14 % in 1512 liver cancers tested; 0.07 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,222 cancer specimens
Comments:
Only 5 deletions, and no insertions or complex mutations are noted on the COSMIC website.