Nomenclature
Short Name:
Obscn
Full Name:
Obscurin-myosin light chain kinase
Alias:
- Obscurin
Classification
Type:
Protein-serine/threonine kinase
Group:
CAMK
Family:
Trio
SubFamily:
NA
Structure
Mol. Mass (Da):
868,484
# Amino Acids:
7968
# mRNA Isoforms:
5
mRNA Isoforms:
924,971 Da (8483 AA; Q5VST9-6); 868,571 Da (7969 AA; Q5VST9-2); 868,484 Da (7968 AA; Q5VST9); 721,547 Da (6620 AA; Q5VST9-3); 427,046 Da (3911 AA; Q5VST9-5)
4D Structure:
Isoform 3 interacts with TTN/titin, ANK1 isoform Mu17/ank1.5, and with calmodulin in a Ca2+-independent manner. Associates with fast skeletal muscle myosin
1D Structure:
Subfamily Alignment
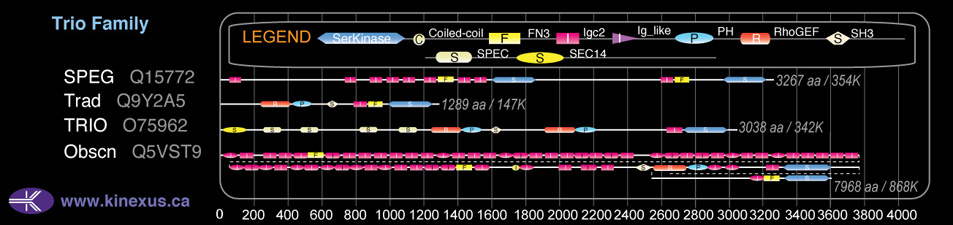
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K272, K3700, K7438.
Serine phosphorylated:
S136, S238, S241, S258, S676, S757, S790, S1062, S2156, S2326, S2443, S2511, S2851, S2889, S2975, S3073, S3099, S3343, S3373, S3528, S3837, S3847, S4076, S4239, S4240, S4750, S4757, S4759, S4787, S4804, S4805, S4871, S4997, S5243, S5244, S5252, S5563, S5669, S5944, S5954, S5955, S6162, S6762, S6773, S6808, S6812, S6817, S6829, S6831, S6881, S6942, S7178, S7232, S7234, S7244, S7487.
Threonine phosphorylated:
T246, T269, T2155, T2776, T2932, T2936, T3064, T3290, T3527, T3530, T3532, T3603, T3620, T3642, T3663, T3708, T3751, T4754, T4788, T4803, T5250, T5254, T5569, T6222, T6971, T7473, T7495.
Tyrosine phosphorylated:
Y65, Y80, Y287, Y3060, Y3342, Y5864, Y6554, Y7705.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 20
20
676
34
1013
 5
5
154
21
459
 0.2
0.2
8
1
0
 4
4
137
99
354
 11
11
375
42
389
 0.3
0.3
9
55
9
 0.2
0.2
8
44
7
 15
15
527
28
762
 5
5
186
19
308
 2
2
55
94
101
 1.3
1.3
43
25
104
 19
19
649
99
592
 0.7
0.7
25
14
48
 3
3
93
17
299
 2
2
72
22
157
 2
2
60
19
143
 0.6
0.6
19
111
70
 0.8
0.8
28
17
54
 18
18
629
105
608
 10
10
349
123
332
 2
2
58
21
113
 1
1
35
21
87
 0.3
0.3
9
2
1
 4
4
132
19
230
 2
2
85
21
173
 17
17
576
65
571
 0.8
0.8
28
26
74
 1
1
35
17
78
 1.2
1.2
41
17
104
 6
6
200
42
176
 25
25
849
18
617
 100
100
3422
39
6256
 7
7
241
127
648
 20
20
688
78
600
 0.8
0.8
29
57
27
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 21.5
21.5
22.5
79 -
-
-
- -
-
-
- -
-
-
- 40.4
40.4
46.1
- -
-
-
- 71.5
71.5
78.1
59 74.1
74.1
81.4
65 -
-
-
- -
-
-
- 47.9
47.9
62.5
45 -
-
-
39 26.4
26.4
40.4
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
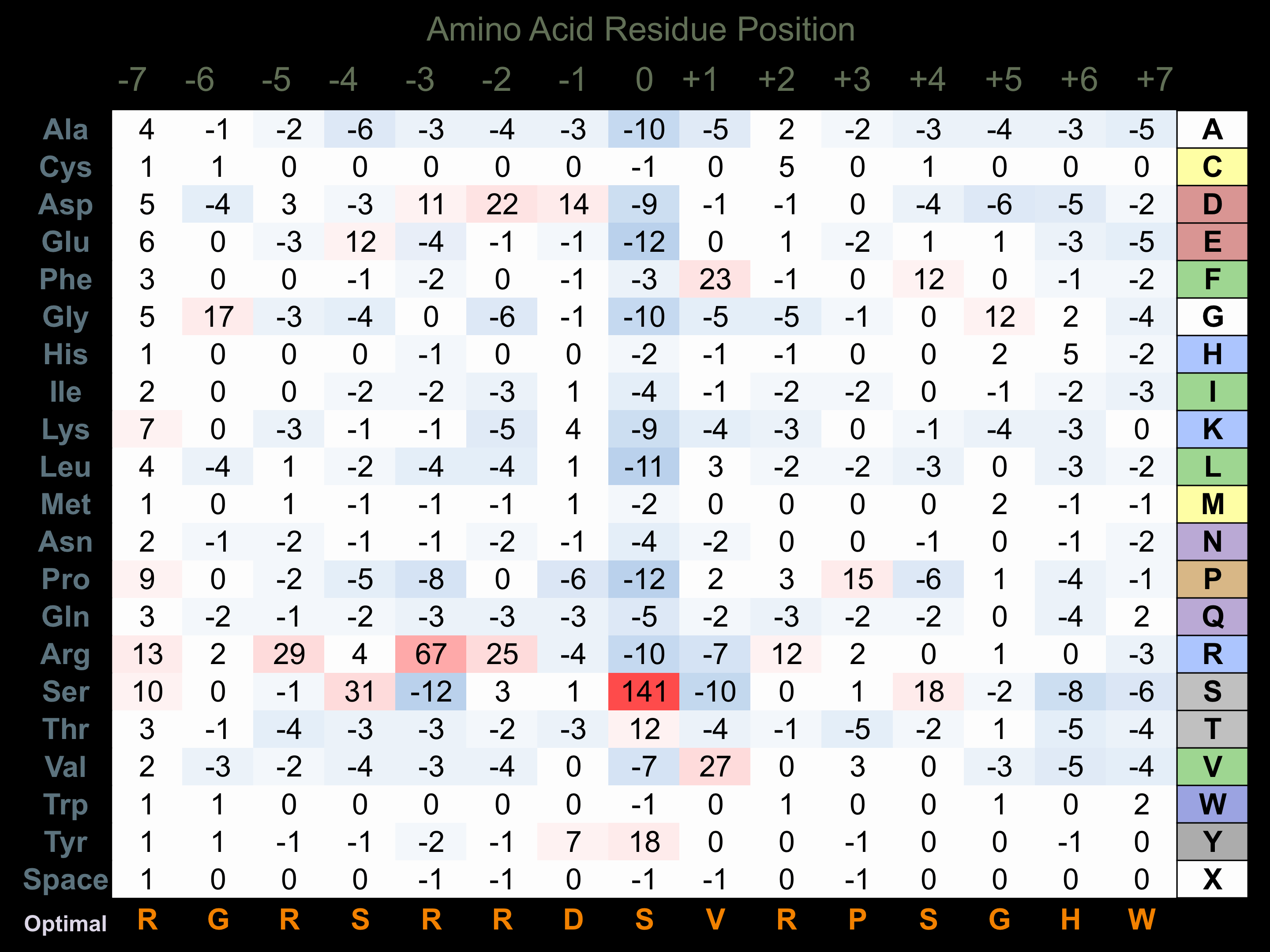
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
2
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Wilm's tumours
Comments:
A chromosomal aberration involving Obscn has been reported in Wilms tumour, and the gene has a translocation t(1;7)(q42;p15) with PTHB1. Due to its large size, there is a greater chance of mutations being detected in this protein in human cancers, but these may be mainly bystander mutations.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in human Breast epithelial hyperplastic enlarged lobular units (HELU) (%CFC= -46, p<0.075).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.04 % in 24440 diverse cancer specimens. This rate is -40 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.23 % in 1178 large intestine cancers tested; 0.2 % in 805 skin cancers tested; 0.2 % in 589 stomach cancers tested; 0.11 % in 602 endometrium cancers tested; 0.11 % in 1619 lung cancers tested; 0.07 % in 605 oesophagus cancers tested; 0.06 % in 500 urinary tract cancers tested; 0.05 % in 229 bone cancers tested; 0.05 % in 1329 breast cancers tested; 0.05 % in 1270 liver cancers tested; 0.05 % in 1187 pancreas cancers tested.
Frequency of Mutated Sites:
None > 8 in 20,139 cancer specimens
Comments:
Over 40 deletion mutations (12 at A7575fs*79), 8 insertions and 4 complex mutations are noted on the COSMIC website.

