Nomenclature
Short Name:
ROS
Full Name:
Proto-oncogene tyrosine-protein kinase ROS1
Alias:
- C-ros oncogene 1 , receptor tyrosine kinase
- C-ros-1
- ROS1
- V-ros UR2 sarcoma virus oncogene homolog 1 (avian)
- V-ros UR2 sarcoma virus oncogene homologue 1 (avian)
- EC 2.7.10.1
- KROS
- MCF3
- Proto-oncogene tyrosine-protein kinase ROS
Classification
Type:
Protein-tyrosine kinase
Group:
TK
Family:
Sev
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
263915
# Amino Acids:
2347
# mRNA Isoforms:
1
mRNA Isoforms:
263,915 Da (2347 AA; P08922)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
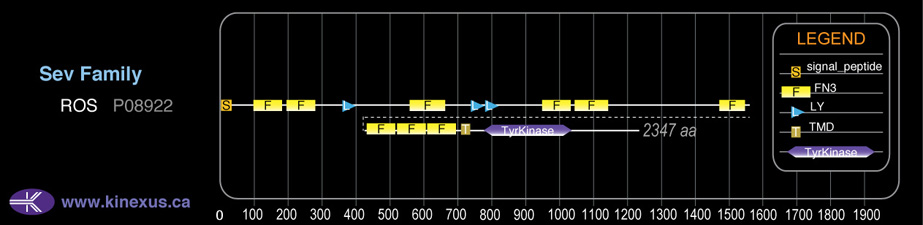
Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K1259, K1504, K1749, K1944.
N-GlcNAcylated:
N52, N114, N123, N324, N352, N396, N471, N607, N628, N706, N714, N732, N939, N961, N1015, N1087, N1090, N1095, N1211, N1272, N1330, N1458, N1461, N1474, N1499, N1565, N1669, N1715, N1738, N1808.
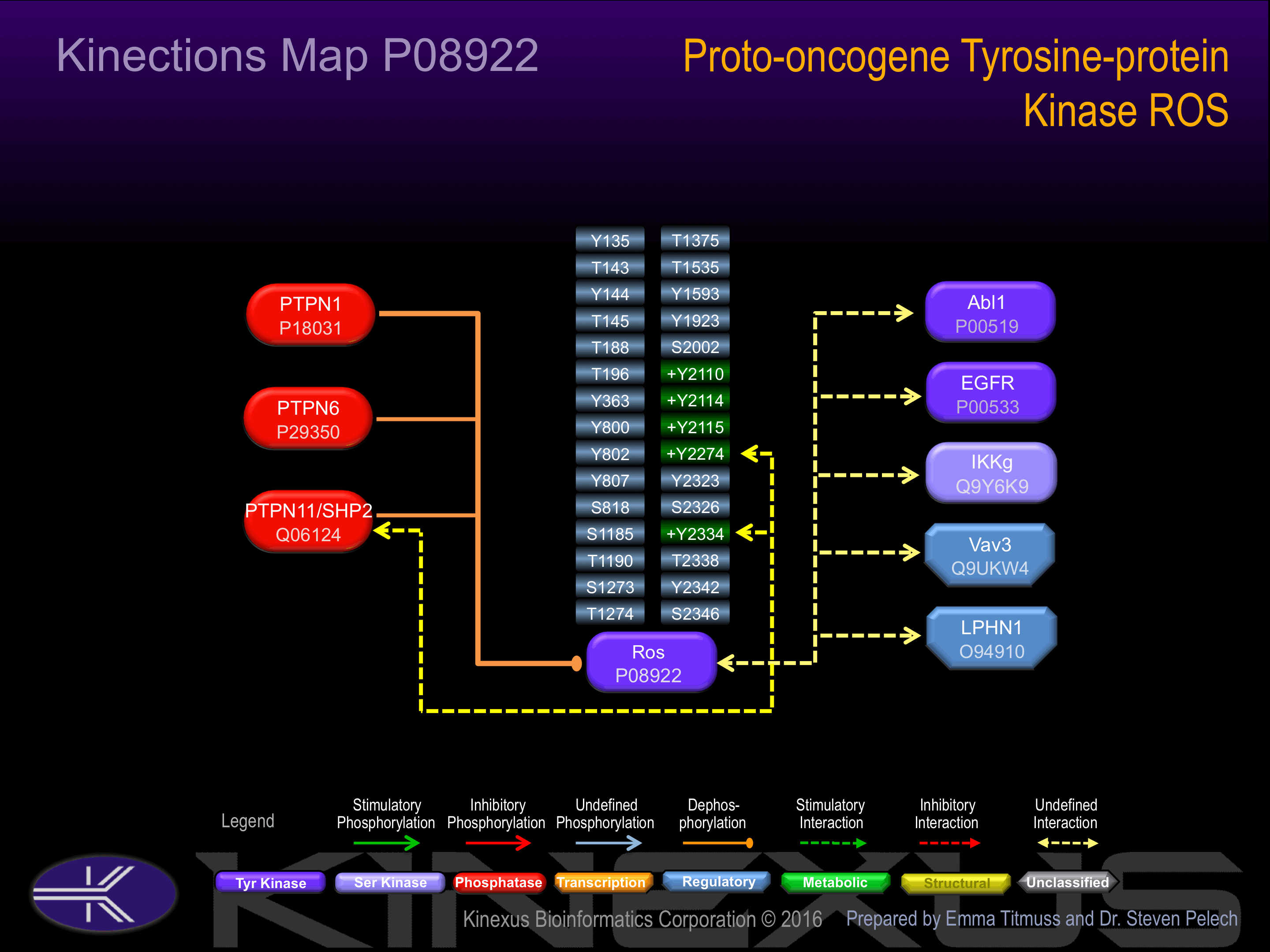
Serine phosphorylated:
S818, S1185, S1273, S2002, S2326, S2346.
Threonine phosphorylated:
T143, T145, T188, T196, T1190, T1274, T1375, T1927, T1930, T2338.
Tyrosine phosphorylated:
Y135, Y144, Y800, Y802, Y807, Y1535, Y1923, Y2023, Y2069, Y2110+, Y2114+, Y2115+, Y2173, Y2227, Y2274, Y2323, Y2334, Y2342.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 18
18
661
22
881
 0.9
0.9
33
9
39
 0.6
0.6
21
1
0
 27
27
960
82
2503
 9
9
331
24
272
 2
2
55
52
144
 3
3
112
29
270
 100
100
3604
25
5452
 7
7
254
10
253
 1
1
54
69
63
 0.7
0.7
26
13
29
 16
16
582
100
618
 0.7
0.7
24
12
11
 0.8
0.8
27
8
22
 0.5
0.5
18
9
19
 0.3
0.3
11
13
9
 3
3
93
105
873
 1
1
51
8
54
 1
1
47
61
28
 8
8
295
84
265
 0.6
0.6
22
10
25
 0.5
0.5
19
11
14
 0.9
0.9
32
10
24
 1
1
35
8
45
 0.4
0.4
15
10
15
 75
75
2717
52
4492
 0.6
0.6
23
15
17
 0.7
0.7
25
7
27
 0.6
0.6
21
6
21
 4
4
132
28
91
 11
11
397
18
284
 27
27
960
26
2223
 2
2
63
51
404
 16
16
589
57
561
 5
5
169
35
113
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.2
99.2
99.5
99 96.6
96.6
98.3
97 -
-
-
85 -
-
-
- 73.4
73.4
79.8
84 -
-
-
- 79.9
79.9
88.3
80 79.8
79.8
87.9
80.5 -
-
-
- 33.7
33.7
47.7
- 53.3
53.3
68.7
55 21.1
21.1
34.9
47 -
-
-
45 -
-
-
- 23.1
23.1
40
32 26.9
26.9
45.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PTPN6 - P29350 |
| 2 | EGFR - P00533 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
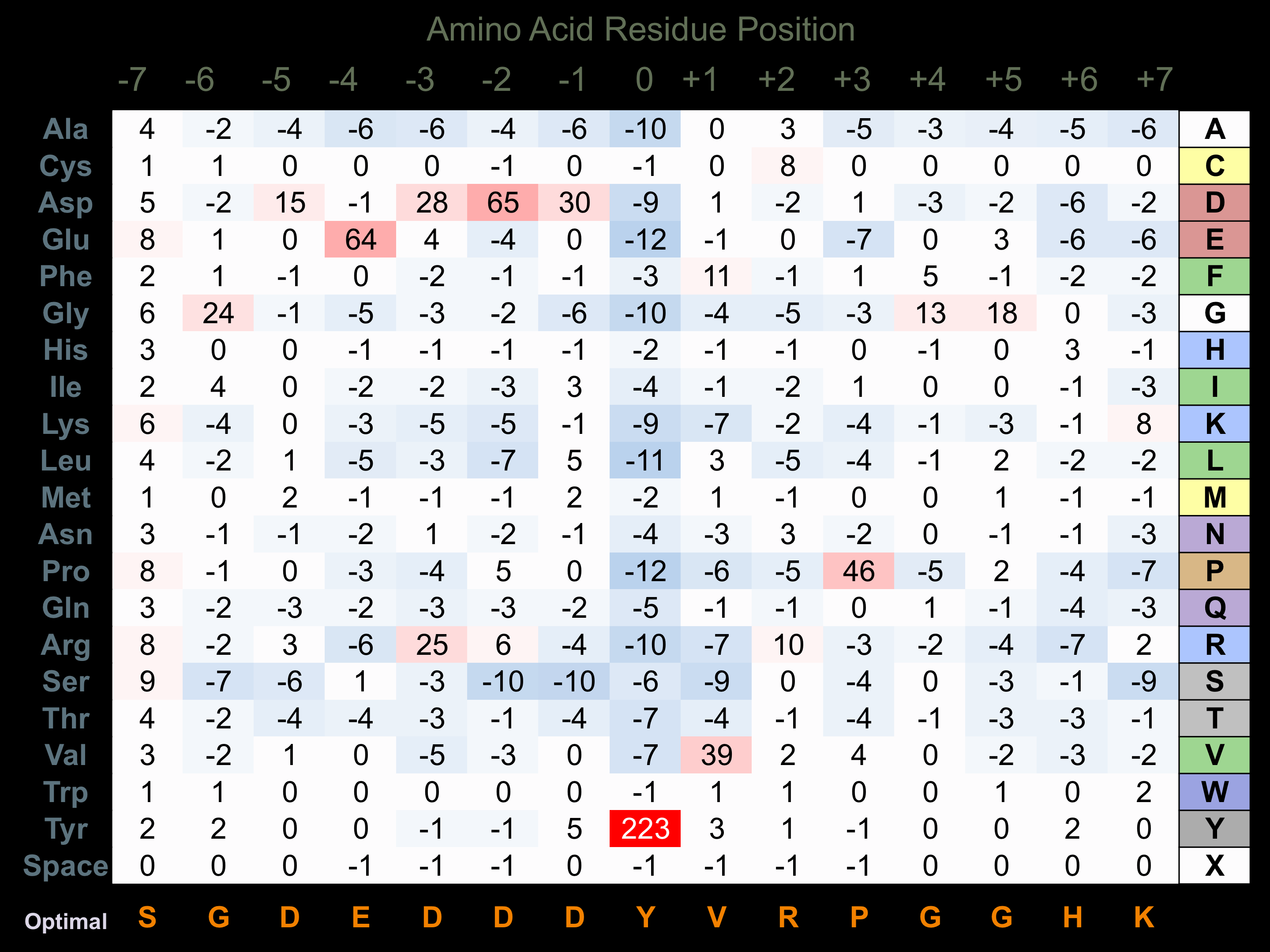
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Glioblastoma multiforme (GBM); Non-small cell lung cancer (NSCLC)
Comments:
ROS1 appears to be an oncoprotein (OP). Cancer-related mutations in human tumours point to a gain of function of the protein kinase. The active form of the protein kinase normally acts to promote tumour cell proliferation. ROS is linked to Glioblastoma Multiforme, in which this protein-tyrosine kinase can be constitutively active. This activity can arise either from ROS fusion with the FIG protein or a deletion (q21q21) leading to GOPC-ROS1. Glioblastoma Multiforme is related to astrocytoma and anaplastic astrocytoma, with regions of anaplastic cells surrounding necrotizing tissue. Glioblastoma Multiforme affects brain, endothelial, and bone tissues. Non-small cell lung carcinoma can arise from a SLC34A2-ROS1 fusion expressing the chimeric CD74-ROS1 protein. In non-small cell lung cancer a gain of function has been observed with MLK1 (MAP3K9), activating ERK downstream.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Uterine leiomyosarcomas (%CFC= +101, p<0.057). The COSMIC website notes an up-regulated expression score for ROS1 in diverse human cancers of 234, which is 0.5-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 1 for this protein kinase in human cancers was 98% lower than the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. A K1980M mutation can lead to impaired phosphotransferase activity, while the mutations Y2274F or Y2334F can lead to loss of phosphorylation and reduced complex formation with PTPN11.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 25640 diverse cancer specimens. This rate is only 24 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.65 % in 805 skin cancers tested; 0.32 % in 1243 large intestine cancers tested; 0.22 % in 629 stomach cancers tested; 0.17 % in 602 endometrium cancers tested; 0.15 % in 1756 lung cancers tested; 0.13 % in 500 urinary tract cancers tested; 0.07 % in 950 upper aerodigestive tract cancers tested; 0.07 % in 904 ovary cancers tested; 0.07 % in 605 oesophagus cancers tested; 0.06 % in 1466 breast cancers tested; 0.06 % in 1314 kidney cancers tested; 0.04 % in 1962 central nervous system cancers tested; 0.04 % in 1942 haematopoietic and lymphoid cancers tested; 0.04 % in 1270 liver cancers tested.
Frequency of Mutated Sites:
None > 5 in 20,733 cancer specimens
Comments:
Broad distribution of mutation sites with many point mutations, deletions, and insertions noted on the COSMIC website.