Nomenclature
Short Name:
CK2a2
Full Name:
Casein kinase II, alpha' chain
Alias:
- Casein kinase 2, alpha prime polypeptide
- Casein kinase II, alpha\' chain
- CSNK2A2
- EC 2.7.1.37
- Kinase CK2-alpha2
- CK II
- CK2A2
- CK2-alpha2
- CSK22
Classification
Type:
Protein-serine/threonine kinase
Group:
Other
Family:
CK2
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
41,213
# Amino Acids:
350
# mRNA Isoforms:
1
mRNA Isoforms:
41,213 Da (350 AA; P19784)
4D Structure:
Tetramer composed of an alpha chain, an alpha' and two beta chains. Also component of a CK2-SPT16-SSRP1 complex composed of SSRP1, SUPT16H, CSNK2A1, CSNK2A2 and CSNK2B, the complex associating following UV irradiation.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 40 | 325 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K50, K97 (N6).
Serine phosphorylated:
S18, S21, S195+, S288, S333, S343.
Tyrosine phosphorylated:
Y13, Y24, Y116, Y210, Y240, Y256.
Ubiquitinated:
K260.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 42
42
1045
22
883
 4
4
89
11
14
 31
31
762
12
522
 21
21
511
94
872
 36
36
898
24
683
 7
7
170
56
145
 12
12
287
29
499
 36
36
902
46
1623
 14
14
337
10
274
 7
7
185
79
173
 7
7
177
35
224
 24
24
593
132
617
 14
14
340
23
350
 4
4
108
9
53
 11
11
283
31
340
 5
5
129
13
27
 6
6
147
441
1170
 17
17
415
19
350
 10
10
245
72
268
 42
42
1044
84
707
 9
9
228
31
299
 13
13
311
33
451
 19
19
462
21
458
 100
100
2478
19
2170
 13
13
316
31
395
 38
38
953
58
967
 12
12
300
26
333
 12
12
293
19
249
 25
25
608
19
581
 3
3
80
28
53
 48
48
1183
18
801
 20
20
486
26
627
 29
29
720
68
994
 38
38
940
57
771
 43
43
1075
35
1177
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 91.7
91.7
92
99 57.9
57.9
58.9
99 -
-
-
99 -
-
-
98 91.4
91.4
92.8
99 -
-
-
- 98.8
98.8
99.7
99 75.1
75.1
82
98 -
-
-
- 98.2
98.2
99.4
- 97.1
97.1
98.5
97 75
75
82.1
- 23.7
23.7
47.1
86 -
-
-
- 77.4
77.4
86
89 79.4
79.4
89.1
- 75
75
86.1
79 71.3
71.3
79.4
- -
-
-
- 69.7
69.7
82.2
- -
-
-
73.5 67.4
67.4
81.4
72 56
56
70.2
67 65.4
65.4
79.7
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | LY6G5B - Q8NDX9 |
| 2 | CSNK2B - P67870 |
| 3 | CSNK2B - P67870 |
| 4 | HSP90AA1 - P07900 |
| 5 | HSP90AA2 - Q14568 |
| 6 | HSP90B1 - P14625 |
| 7 | HDAC2 - Q92769 |
| 8 | HDAC1 - Q13547 |
| 9 | EEF1B2 - P24534 |
| 10 | CAV1 - Q03135 |
| 11 | CALM1 - P62158 |
| 12 | CSN3 - P07498 |
| 13 | HNRNPC - P07910 |
| 14 | CLTB - P09497 |
| 15 | PPP1R1B - Q9UD71 |
Regulation
Activation:
Activated by the CK2-beta regulatory subunit. Interaction with p38-alpha MAP kinase induces stimulation of CK2's catalytic activity.
Inhibition:
CK2 is potently inhibited by heparin.
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| Bamacan | Q9UQE7 | S1067 | GDVEGSQSQDEGEGS | |
| CK2-B | Q5SRQ6 | S21 | TSADVKMSSSEEVSW | + |
| CK2-B | Q5SRQ6 | S22 | SADVKMSSSEEVSWI | |
| CTNNB1 | P35222 | S29 | VSHWQQQSYLDSGIH | |
| CTNNB1 | P35222 | T102 | RAAMFPETLDEGMQI | |
| CTNNB1 | P35222 | T112 | EGMQIPSTQFDAAHP | |
| Cyclin H (CCNH) | P51946 | T315 | KHEEEEWTDDDLVES | |
| Cyclin H (CCNH) | P51946 | Y99 | LNNSVMEYHPRIIML | |
| HSF1 | Q00613 | T142 | DSVTKLLTDVQLMKG | + |
| NKX3-1 | Q99801 | T89 | AAPEEAETLAETEPE | + |
| NKX3-1 | Q99801 | T93 | EAETLAETEPERHLG | + |
| SRPK1 | Q96SB4 | S222 | QEEEILGSDDDEQED | + |
| SRPK1 | Q96SB4 | S726 | DYLFEPHSGEEYTRD | + |
| SRPK2 | P78362 | S52 | PEEEILGSDDEEQED | + |
| SRPK2 | P78362 | S588 | DYLFEPHSGEDYSRD | + |
| XRCC1 | P18887 | S485 | QDNGAEDSGDTEDEL | |
| XRCC1 | P18887 | S518 | GEDPYAGSTDENTDS | |
| XRCC1 | P18887 | T488 | GAEDSGDTEDELRRV | |
| XRCC1 | P18887 | T519 | EDPYAGSTDENTDSE | |
| XRCC1 | P18887 | T523 | AGSTDENTDSEEHQE |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
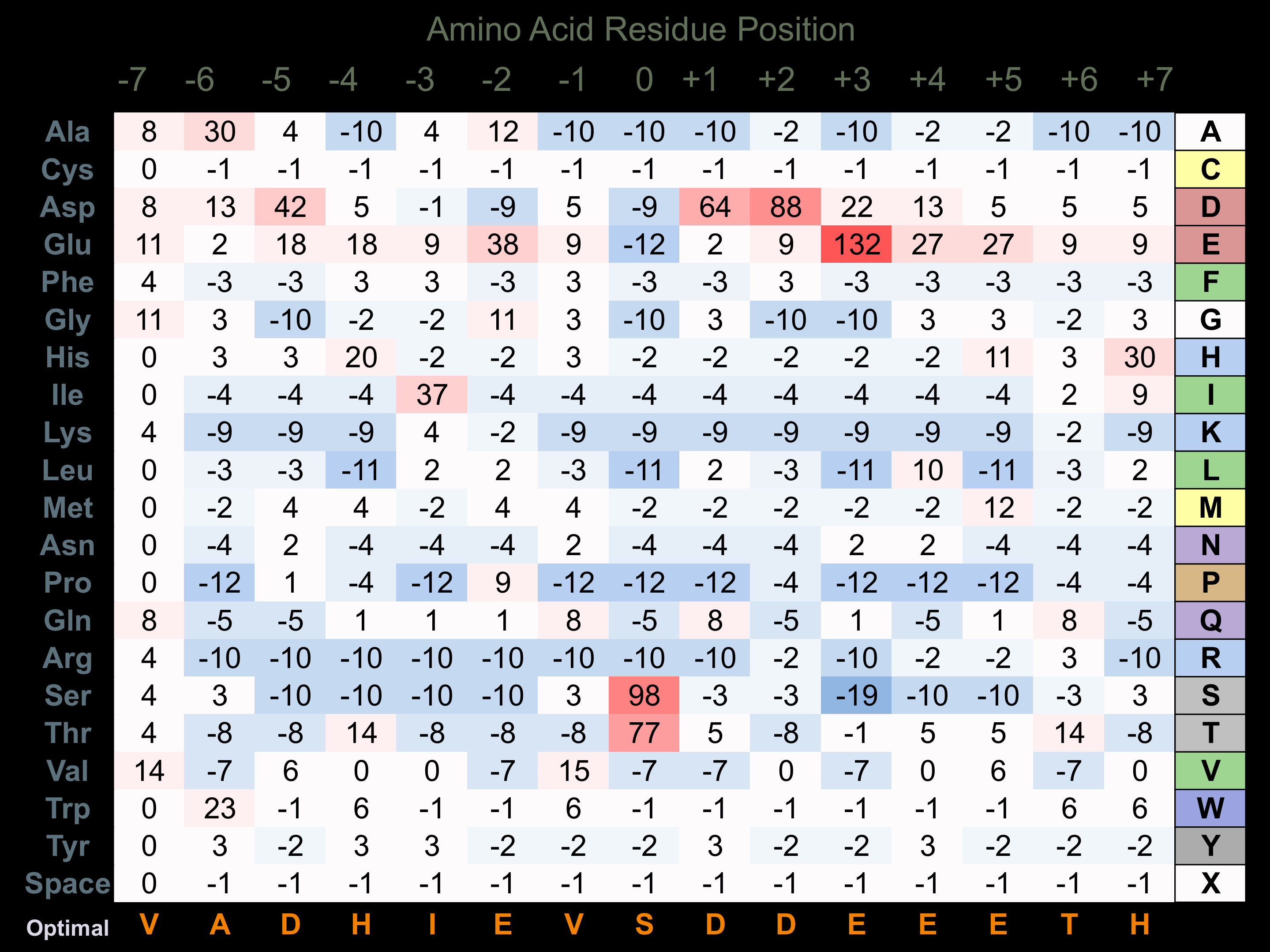
Matrix Type:
Experimentally derived from alignment of 21 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Reproductive disorders
Specific Diseases (Non-cancerous):
Globozoospermia; PANDAS; Theileriasis
Comments:
In animal studies, disruption of CSNK2A2 gene expression in mice leads to infertility associated with oligospermia and globozoospermia phenotypes that closely resemble human globozoospermia. Interestingly, mutation of ORB5, the ortholog of CSNK2A2 in fission yeast, produces a spherical phenotype similar to the spherical sperm heads observed in globozoospermia, indicating a potential influnce of this gene on cytoskeletal integrity. Therefore, mutations in the CSNK2A2 gene may be a cause of inherited abnormalities in sperm morphogenesis, and more specifically they are implicated in the pathogenesis of globozoospermia. Globozoospermia is a rare (incidence
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +78, p<0.001); Clear cell renal cell carcinomas (cRCC) (%CFC= +64, p<0.001); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +249, p<0.0006); Large B-cell lymphomas (%CFC= +92, p<0.012); and Skin fibrosarcomas (%CFC= -57).
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 24433 diverse cancer specimens. This rate is only -27 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 1270 large intestine cancers tested; 0.19 % in 603 endometrium cancers tested; 0.13 % in 864 skin cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R230G (3).
Comments:
Only 1 insertion, and no deletions or complex mutations are noted on the COSMIC website.

