Nomenclature
Short Name:
ANPb
Full Name:
Atrial natriuretic peptide receptor B
Alias:
- ANP-B
- EC 4.6.1.2
- GC-B
- Guanylate cyclase
- NPR2
- NPRB; GUCY2B; Natriuretic peptide receptor B/guanylate cyclase B;
- ANPRB
- Atrial natriuretic peptide B-type receptor
- Atrial natriuretic peptide receptor B precursor
- Atrionatriuretic peptide receptor B
Classification
Type:
Protein-serine/threonine kinase
Group:
RGC
Family:
RGC
SubFamily:
NA
Structure
Mol. Mass (Da):
117,022
# Amino Acids:
1047
# mRNA Isoforms:
2
mRNA Isoforms:
117,022 Da (1047 AA; P20594); 111,208 Da (995 AA; P20594-2)
4D Structure:
NA
1D Structure:
Subfamily Alignment
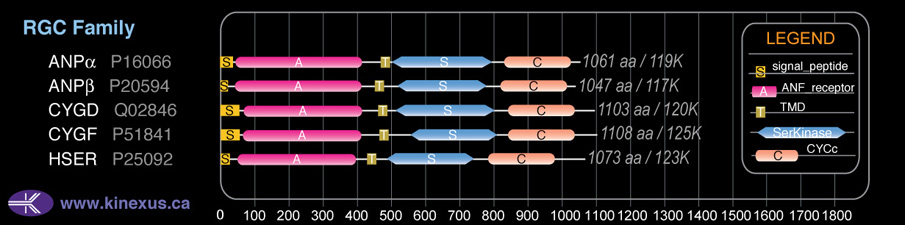
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 1 | 16 | signal_peptide |
| 44 | 401 | ANF_receptor |
| 455 | 475 | TMD |
| 512 | 802 | Pkinase |
| 825 | 1019 | CYCc |
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S489, S513, S518, S523, S526.
Threonine phosphorylated:
T516, T529, T820.
Tyrosine phosphorylated:
Y747, Y808.
N-GlcNAcylated:
N24, N35, N161, N195, N244, N277, N349.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
798
29
923
 6
6
47
15
31
 26
26
205
23
209
 36
36
289
96
467
 98
98
780
23
630
 7
7
57
82
136
 26
26
208
31
491
 66
66
524
51
769
 49
49
393
17
360
 10
10
79
94
88
 12
12
94
41
137
 74
74
587
183
606
 9
9
75
45
87
 6
6
51
12
39
 7
7
57
33
66
 7
7
59
15
32
 8
8
60
129
63
 7
7
57
30
60
 10
10
80
98
78
 71
71
565
109
585
 16
16
126
34
120
 10
10
83
38
95
 12
12
94
33
99
 11
11
87
32
94
 10
10
76
30
86
 77
77
616
65
909
 7
7
54
48
70
 11
11
90
32
91
 37
37
296
32
345
 14
14
111
28
76
 3
3
27
12
4
 91
91
727
36
867
 1
1
8
34
7
 82
82
651
57
623
 11
11
84
35
85
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
99.9
100 99.8
99.8
100
100 -
-
-
98 -
-
-
99 89.1
89.1
90.1
99 -
-
-
- 98.6
98.6
99.4
99 98.6
98.6
99.3
99 -
-
-
- 64.6
64.6
67
- -
-
-
80 72.3
72.3
83
- 58.2
58.2
69.8
- -
-
-
- 32.5
32.5
49.1
48 -
-
-
- 28.8
28.8
46.8
48 36.3
36.3
52.3
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Regulation
Activation:
Activated by binding of atrial natriuretic peptide (ANP), which activates the guanylate cyclase activity of this receptor. Phosphorylation at Ser-513, Ser-518, Ser-523, Ser-526 and Thr-516 sensitize NPR2 for binding ANP.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
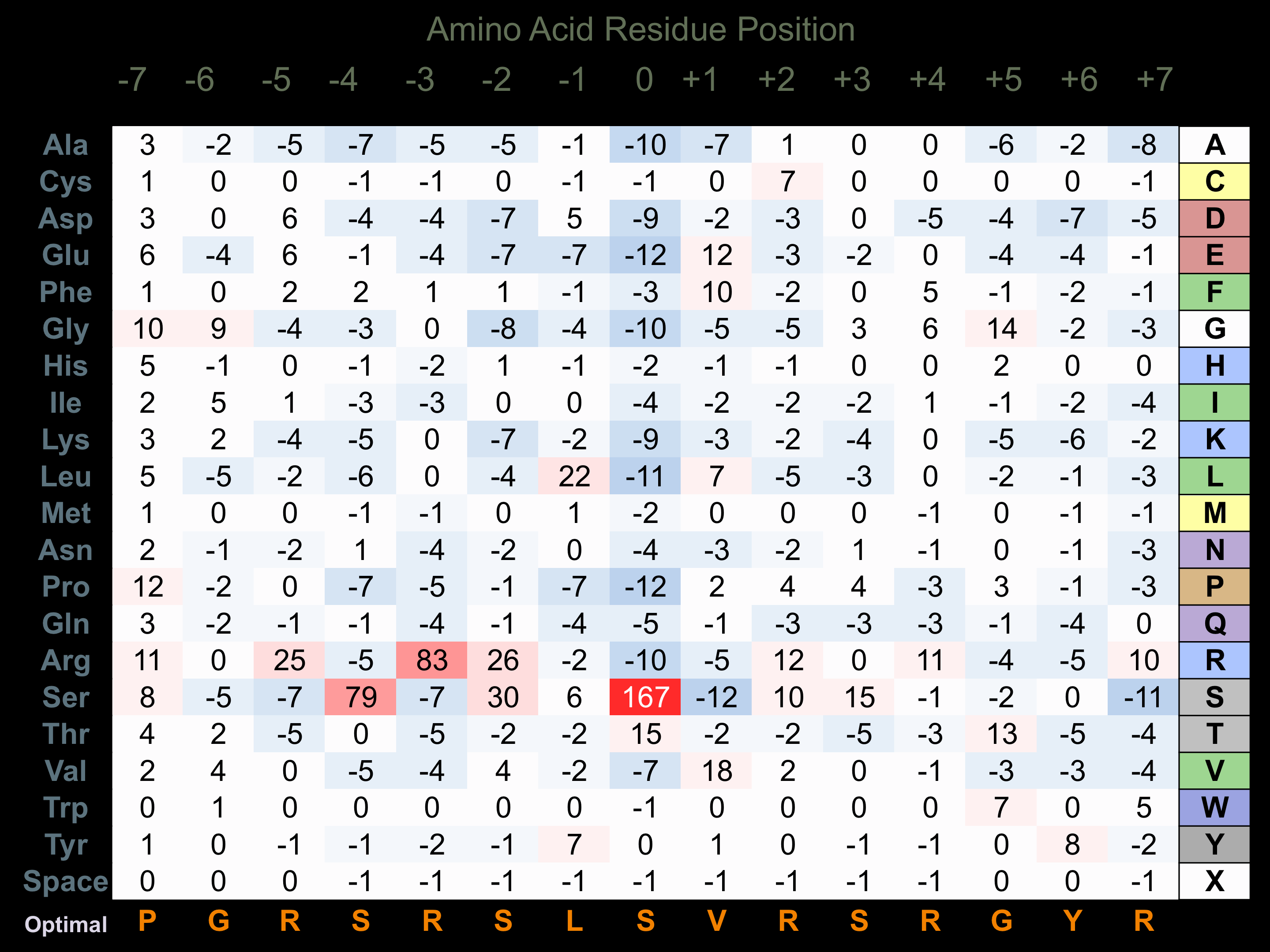
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Disease Linkage
General Disease Association:
Dwarfism and hypertension
Specific Diseases (Non-cancerous):
Acromesomelic dysplasia, Maroteaux Type; Acromesomelic dysplasia, Maroteaux Type; Acromesomelic dysplasia; Short stature; Essential hypertension; Aortic valve stenosis; Renovascular hypertension; Tall stature - scoliosis - Macrodactyly of the great toes
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Pituitary adenomas (ACTH-secreting) (%CFC= -52, p<0.038); Prostate cancer - primary (%CFC= +67, p<0.021); and Skin melanomas (%CFC= -55, p<0.076). The COSMIC website notes an up-regulated expression score for ANPb in diverse human cancers of 474, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 26 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis. W115G, T297M and G413E mutations can all reduce catalytic activity.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.07 % in 24730 diverse cancer specimens. This rate is only -5 % lower and is very similar to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.4 % in 864 skin cancers tested; 0.28 % in 589 stomach cancers tested; 0.26 % in 1270 large intestine cancers tested; 0.24 % in 603 endometrium cancers tested; 0.17 % in 550 urinary tract cancers tested; 0.05 % in 942 upper aerodigestive tract cancers tested.
Frequency of Mutated Sites:
None > 9 in 20,013 cancer specimens
Comments:
Only 2 deletions, and no insertions or complex mutations are noted on the COSMIC website.

