Nomenclature
Short Name:
CDK8
Full Name:
Cyclin-dependent kinase 8
Alias:
- EC 2.7.11.22
- EC 2.7.11.23
- Kinase Cdk8
- Protein kinase K35
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDK8
Specific Links
Structure
Mol. Mass (Da):
53,284
# Amino Acids:
464
# mRNA Isoforms:
2
mRNA Isoforms:
53,284 Da (464 AA; P49336); 53,156 Da (463 AA; P49336-2)
4D Structure:
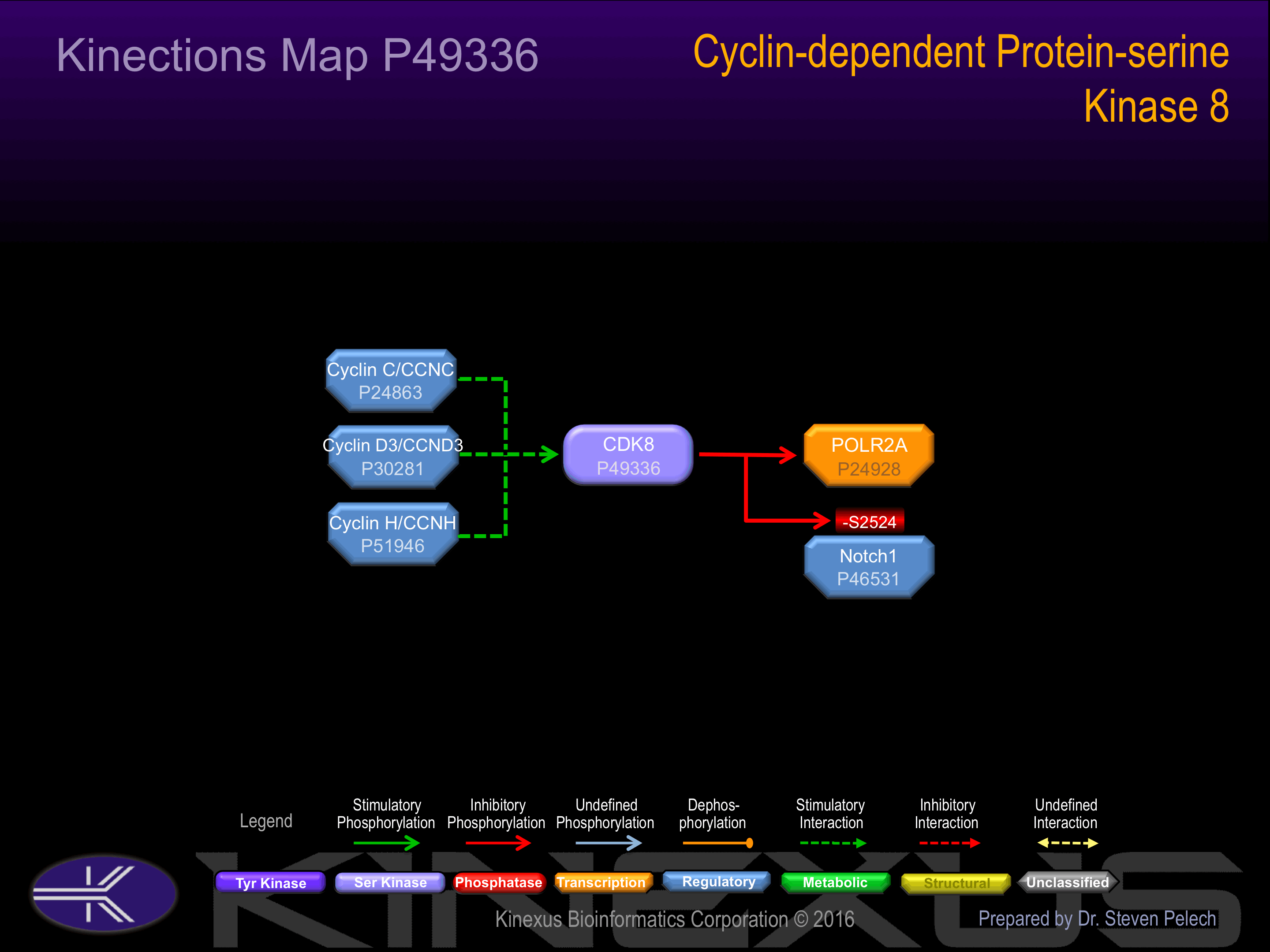
Component of the Mediator complex, which is composed of MED1, MED4, MED6, MED7, MED8, MED9, MED10, MED11, MED12, MED13, MED13L, MED14, MED15, MED16, MED17, MED18, MED19, MED20, MED21, MED22, MED23, MED24, MED25, MED26, MED27, MED29, MED30, MED31, CCNC, CDK8 and CDC2L6/CDK11. The MED12, MED13, CCNC and CDK8 subunits form a distinct module termed the CDK8 module. Mediator containing the CDK8 module is less active than Mediator lacking this module in supporting transcriptional activation. Individual preparations of the Mediator complex lacking one or more distinct subunits have been variously termed ARC, CRSP, DRIP, PC2, SMCC and TRAP. The cylin/CDK pair formed by CCNC/CDK8 also associates with the large subunit of RNA polymerase II. Interacts with CTNNB1, GLI3 and MAML1.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 21 | 335 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K402.
Serine phosphorylated:
S11, S80, S130, S413.
Tyrosine phosphorylated:
Y133, Y141.
Ubiquitinated:
K52.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 100
100
1297
28
995
 3
3
36
14
28
 71
71
923
13
1270
 20
20
253
104
341
 70
70
913
28
516
 3
3
38
55
21
 23
23
299
43
473
 29
29
370
33
554
 54
54
696
10
703
 10
10
126
59
182
 16
16
206
30
432
 36
36
467
152
614
 18
18
235
24
454
 4
4
46
12
29
 35
35
459
27
1359
 5
5
60
19
17
 10
10
135
208
617
 24
24
307
22
603
 12
12
155
61
360
 63
63
813
102
594
 25
25
318
26
775
 17
17
219
28
427
 33
33
432
22
722
 43
43
560
22
775
 26
26
338
26
802
 54
54
703
70
598
 18
18
231
27
453
 19
19
251
22
542
 33
33
426
22
771
 5
5
59
14
34
 56
56
727
18
477
 74
74
963
31
2766
 21
21
271
72
766
 59
59
771
83
655
 28
28
361
48
395
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 89.7
89.7
89.7
100 -
-
-
100 -
-
-
99.5 -
-
-
- 99.6
99.6
99.6
100 -
-
-
- 98.9
98.9
99.1
99 26.5
26.5
42.5
99 -
-
-
- 99.3
99.3
99.3
- 99.1
99.1
99.6
98 87.1
87.1
88.8
97 95.7
95.7
97.4
96 -
-
-
- 71.1
71.1
77.8
85 -
-
-
- 47.1
47.1
60.2
63 -
-
-
- -
-
-
- -
-
-
- -
-
-
- 38.5
38.5
55.1
- -
-
-
42 -
-
-
40
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CCNC - P24863 |
| 2 | MED6 - O75586 |
| 3 | MED21 - Q13503 |
| 4 | MED1 - Q15648 |
| 5 | MED14 - O60244 |
| 6 | MED10 - Q9BTT4 |
| 7 | POLR2A - P24928 |
| 8 | MED7 - O43513 |
| 9 | MED16 - Q9Y2X0 |
| 10 | MED13 - Q9UHV7 |
| 11 | MED17 - Q9NVC6 |
| 12 | CDKN2B - P42772 |
| 13 | MAML1 - Q92585 |
| 14 | MED9 - Q9NWA0 |
| 15 | MED20 - Q9H944 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
Recruited through interaction with MAML1 to hyperphosphorylate the intracellular domain of NOTCH, leading to its degradation.
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
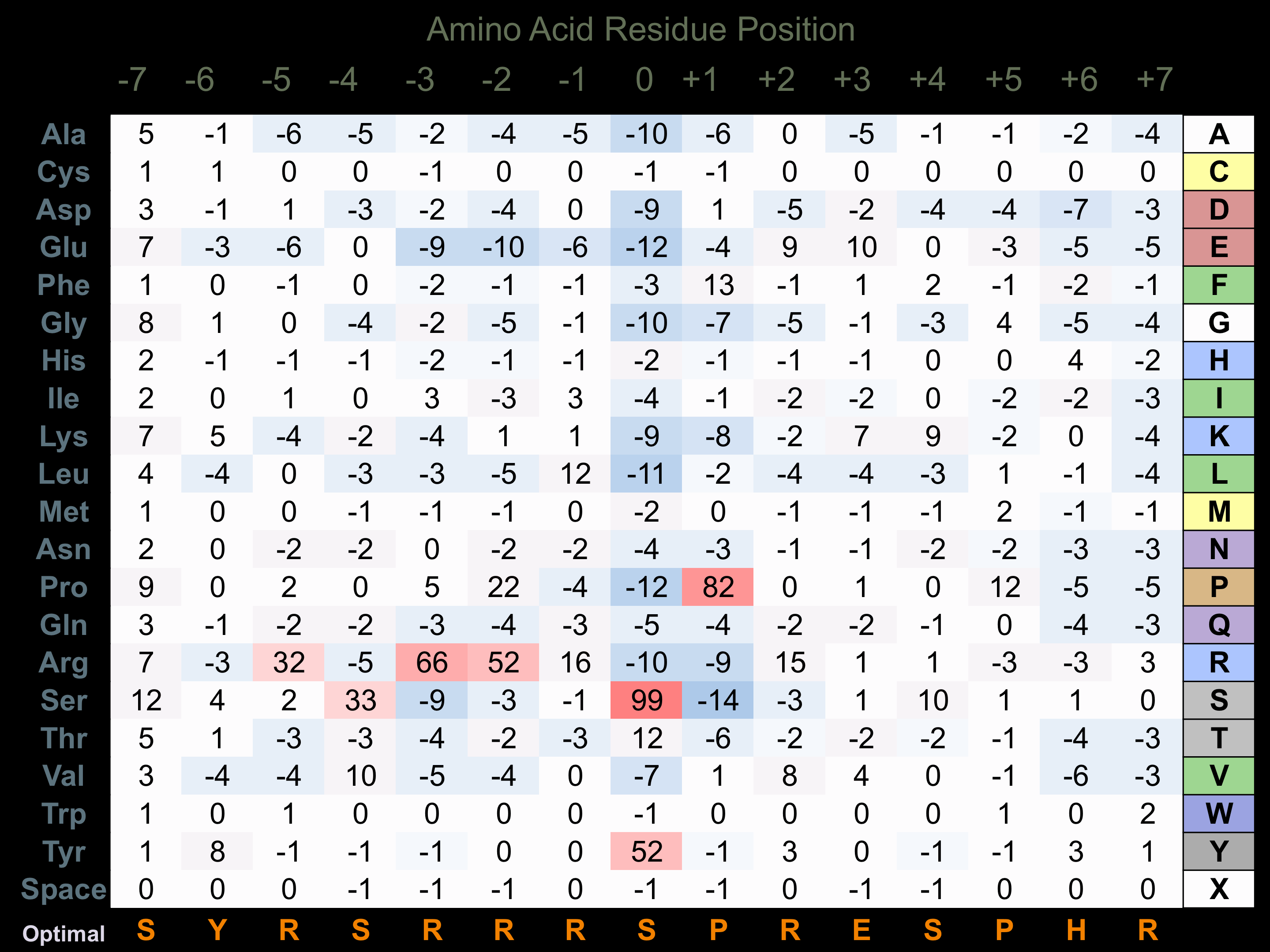
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
| AST-487 | Kd = 1.4 nM | 11409972 | 574738 | 18183025 |
| Linifanib | Kd = 95 nM | 11485656 | 223360 | 18183025 |
| Alvocidib | Kd = 120 nM | 9910986 | 428690 | 18183025 |
| Foretinib | Kd = 130 nM | 42642645 | 1230609 | 22037378 |
| Doramapimod | Kd = 220 nM | 156422 | 103667 | 18183025 |
| Sorafenib | Kd = 310 nM | 216239 | 1336 | 18183025 |
| Staurosporine | Kd = 510 nM | 5279 | 18183025 | |
| WZ3146 | Kd > 1 µM | 44607360 | 20033049 | |
| WZ4002 | Kd > 1 µM | 44607530 | 20033049 | |
| AC1NS7CD | Kd = 1.2 µM | 5329665 | 295136 | 22037378 |
| SNS032 | Kd = 1.2 µM | 3025986 | 296468 | 18183025 |
| PLX4720 | Kd = 1.9 µM | 24180719 | 1230020 | 22037378 |
| CP724714 | Kd = 2.3 µM | 9874913 | 483321 | 18183025 |
| A674563 | Kd = 3.5 µM | 11314340 | 379218 | 22037378 |
| Vatalanib | Kd = 4.5 µM | 151194 | 101253 | 22037378 |
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Colon cancer
Comments:
CDK8 may be a tumour requiring protein (TRP). CDK8 is upregulated in a portion of colon cancers, and inhibition of CDK8 inhibits colon cancer cell division. CDK8 up-regulation can lead to E2F1 inhibition and inhibition of apoptosis.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +65, p<0.01); and Breast epithelial carcinomas (%CFC= +181, p<0.025). The COSMIC website notes an up-regulated expression score for CDK8 in diverse human cancers of 642, which is 1.4-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 112 for this protein kinase in human cancers was 1.9-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis. Kinase activity and transcription repression dependent on TFIIH can be inhibited through a CDK8 D173A mutation.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.08 % in 25101 diverse cancer specimens. This rate is very similar (+ 10% higher) to the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.48 % in 1270 large intestine cancers tested; 0.26 % in 895 skin cancers tested; 0.24 % in 273 cervix cancers tested; 0.21 % in 603 endometrium cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,294 cancer specimens
Comments:
Only 4 deletions, no insertions or complex mutations are noted on the COSMIC website.