Nomenclature
Short Name:
CLK1
Full Name:
Dual specificity protein kinase CLK1
Alias:
- CDC like kinase 1
- CDC-like kinase 1
- CLK
- EC 2.7.1.112
- EC 2.7.12.1
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CLK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
57291
# Amino Acids:
484
# mRNA Isoforms:
3
mRNA Isoforms:
61,757 Da (526 AA; P49759-3); 57,291 Da (484 AA; P49759); 16,570 Da (136 AA; P49759-2)
4D Structure:
Interacts with PPIG.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
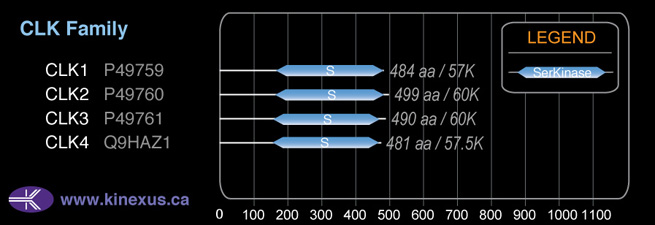
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 161 | 477 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S24, S34, S36, S37, S54, S59, S61, S101, S102, S104, S122, S140, S328, S337+, S341-.
Threonine phosphorylated:
T123, T138+, T330+, T338+, T342-.
Tyrosine phosphorylated:
Y56, Y76, Y80, Y345-.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 10
10
796
16
700
 1.4
1.4
113
12
101
 28
28
2336
13
1832
 12
12
1003
58
1629
 13
13
1043
14
902
 13
13
1086
45
2367
 8
8
673
23
682
 10
10
867
33
964
 10
10
825
10
809
 2
2
139
58
97
 13
13
1084
29
1835
 13
13
1090
130
1519
 33
33
2769
24
3594
 3
3
228
12
145
 17
17
1397
27
2093
 3
3
239
9
88
 3
3
263
197
599
 9
9
781
22
919
 6
6
537
61
879
 11
11
929
56
929
 16
16
1352
26
1614
 25
25
2081
27
2869
 33
33
2756
14
2685
 6
6
511
22
640
 29
29
2428
26
3097
 19
19
1595
38
1339
 15
15
1264
27
1831
 26
26
2133
22
2621
 25
25
2112
22
2668
 0.2
0.2
13
14
12
 13
13
1088
18
209
 100
100
8290
21
12254
 2
2
153
56
243
 12
12
996
26
745
 3
3
222
22
313
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 99.8
99.8
100
99 97.9
97.9
98.8
98 -
-
-
- -
-
-
- 78.7
78.7
86.8
92 -
-
-
- 89
89
93.6
90 50
50
65.9
91 -
-
-
- 86.6
86.6
91.7
- 22.4
22.4
39.2
76 68
68
79.1
71 53.8
53.8
68.3
- -
-
-
- 31.9
31.9
42.7
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 37.8
37.8
56.2
- 27.5
27.5
45
35 -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | PTPN1 - P18031 |
| 2 | PPIG - Q13427 |
| 3 | MBP - P02686 |
| 4 | SFRS6 - Q13247 |
| 5 | LUC7L3 - O95232 |
| 6 | UBL5 - Q9BZL1 |
| 7 | PRPF4 - O43172 |
| 8 | SFRS3 - P84103 |
| 9 | NCOR1 - O75376 |
| 10 | BCLAF1 - Q9NYF8 |
| 11 | SAFB - Q15424 |
| 12 | SLU7 - O95391 |
| 13 | PIAS4 - Q8N2W9 |
| 14 | SRPK2 - P78362 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
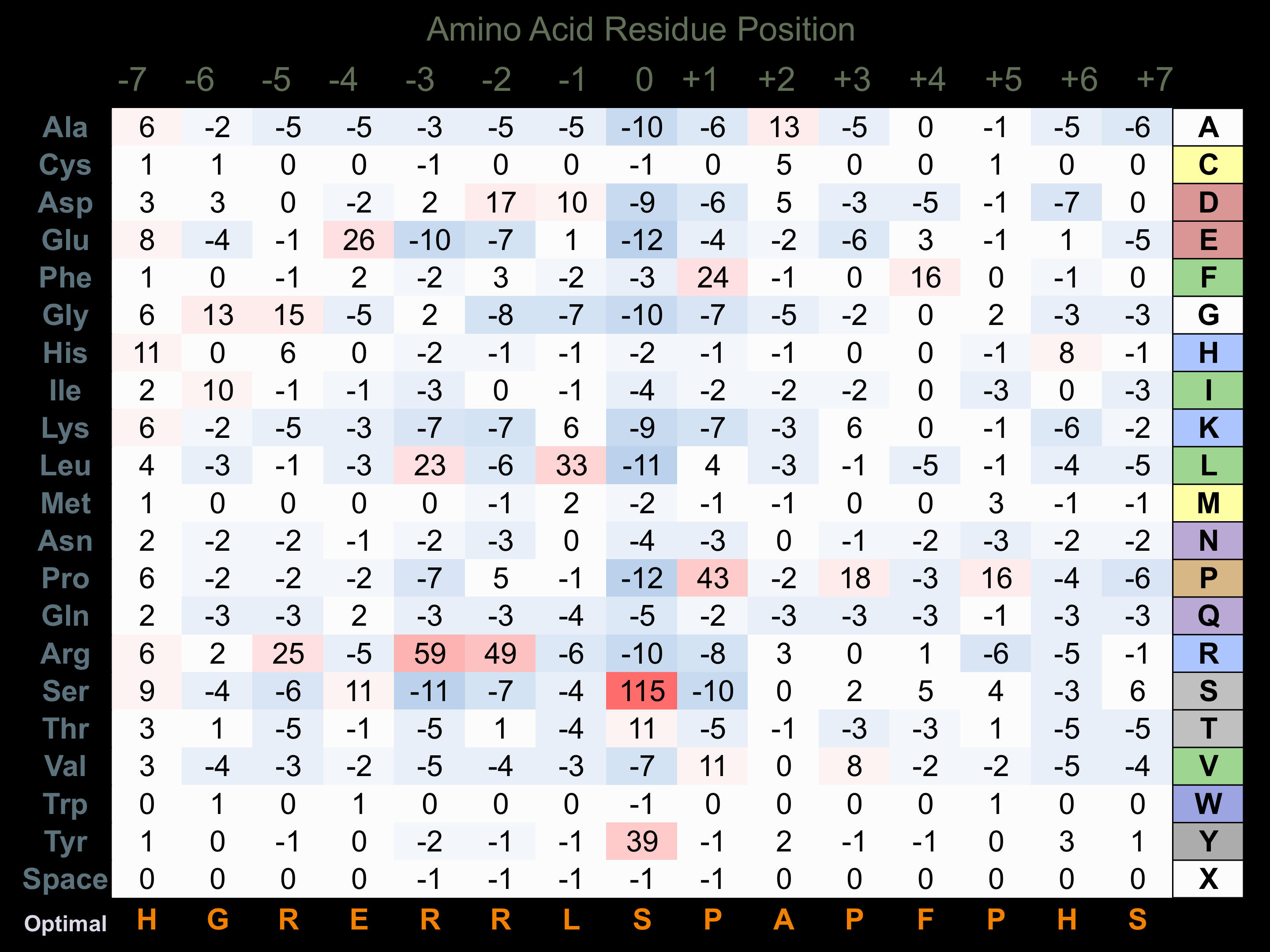
Matrix Type:
Predicted from the application of the Kinexus Kinase Substrate Predictor Version 2.0 algorithm, which was trained with over 10,000 kinase-protein substrate pairs and 8,000 kinase-peptide substrate pairs.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Facial disorders
Specific Diseases (Non-cancerous):
3mc syndrome 2
Comments:
CLK1 regulates the alternative splicing of tissue factor pre-mRNA in endothelial cells and adenovirus E1A pre-mRNA. A small molecule inhibitor of CLK1 significantly reduces influenza virus replication.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Bladder carcinomas (%CFC= +54, p<0.041); Cervical cancer stage 2A (%CFC= +61, p<0.008); Ovary adenocarcinomas (%CFC= -74, p<0.0001); Skin fibrosarcomas (%CFC= -63); and Skin melanomas - malignant (%CFC= -61, p<0.001). The COSMIC website notes an up-regulated expression score for CLK1 in diverse human cancers of 354, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 24 for this protein kinase in human cancers was 0.4-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.09 % in 24751 diverse cancer specimens. This rate is only 17 % higher than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.38 % in 273 cervix cancers tested; 0.34 % in 603 endometrium cancers tested; 0.24 % in 864 skin cancers tested; 0.23 % in 1270 large intestine cancers tested; 0.18 % in 589 stomach cancers tested; 0.15 % in 1512 liver cancers tested; 0.13 % in 1634 lung cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,034 cancer specimens
Comments:
Only 2 deletions, no insertions or complex mutations are noted on the COSMIC website.

