Nomenclature
Short Name:
CDK7
Full Name:
Cyclin-dependent kinase 7
Alias:
- 39 kDa protein kinase
- CAK
- CRK4
- EC 2.7.11.22
- EC 2.7.11.23
- TFIIH basal transcription factor complex kinase subunit
- CAK1
- CDK-activating kinase
- CDKN7
- CR4 protein kinase
Classification
Type:
Protein-serine/threonine kinase
Group:
CMGC
Family:
CDK
SubFamily:
CDK7
Specific Links
Structure
Mol. Mass (Da):
39,038
# Amino Acids:
346
# mRNA Isoforms:
1
mRNA Isoforms:
39,038 Da (346 AA; P50613)
4D Structure:
Associates primarily with cyclin H and MAT1 to form the CAK complex. CAK can further associate with the core-TFIIH to form the TFIIH basal transcription factor. Interacts with PUF60.
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 12 | 295 | Pkinase |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S7, S161, S164-, S321.
Threonine phosphorylated:
T170+, T175-, T287+, T332.
Tyrosine phosphorylated:
Y169+.
Ubiquitinated:
K139, K160, K291, K328.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 25
25
884
16
1064
 1.2
1.2
42
12
21
 2
2
74
1
0
 13
13
467
150
1287
 24
24
854
14
770
 7
7
261
51
353
 14
14
479
23
511
 100
100
3504
30
4802
 17
17
601
10
581
 2
2
87
59
31
 5
5
181
84
608
 13
13
466
117
604
 1.4
1.4
48
12
17
 1.3
1.3
45
12
30
 1.1
1.1
39
13
17
 3
3
92
9
22
 1.1
1.1
40
188
26
 1
1
35
10
24
 8
8
292
148
1249
 22
22
768
56
783
 1.3
1.3
46
14
21
 2
2
54
14
29
 3
3
91
10
27
 9
9
318
102
951
 2
2
65
14
41
 34
34
1204
38
1884
 3
3
108
15
34
 1.3
1.3
47
10
16
 2
2
53
10
24
 0.8
0.8
29
14
25
 31
31
1099
18
742
 15
15
518
21
532
 16
16
560
55
877
 27
27
950
31
794
 36
36
1270
22
3283
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 88.1
88.1
88.1
100 99.7
99.7
99.7
100 -
-
-
97 -
-
-
98 92.6
92.6
95.4
94.5 -
-
-
- 95
95
96.8
93 91
91
92.4
95 -
-
-
- 87.5
87.5
88.7
- 39.8
39.8
56.3
95 85.2
85.2
90.3
- 42.2
42.2
59.5
86 -
-
-
- 65.7
65.7
78.4
67.5 63.8
63.8
75.1
- 39.3
39.3
57.5
61 -
-
-
- -
-
-
- 39.3
39.3
57.8
- -
-
-
58 49.1
49.1
65.2
57 41
41
60.6
49 -
-
-
42
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | POLR2G - P62487 |
| 2 | SP3 - Q02447 |
| 3 | MNAT1 - P51948 |
| 4 | GTF2H1 - P32780 |
| 5 | ESR1 - P03372 |
| 6 | TP53 - P04637 |
| 7 | CDK2 - P24941 |
| 8 | CDC2 - P06493 |
| 9 | ERCC2 - P18074 |
| 10 | PCGF6 - Q9BYE7 |
| 11 | BRCA1 - P38398 |
| 12 | NEK6 - Q9HC98 |
| 13 | RARA - P10276 |
| 14 | THRA - P10827 |
| 15 | MTA1 - Q13330 |
Regulation
Activation:
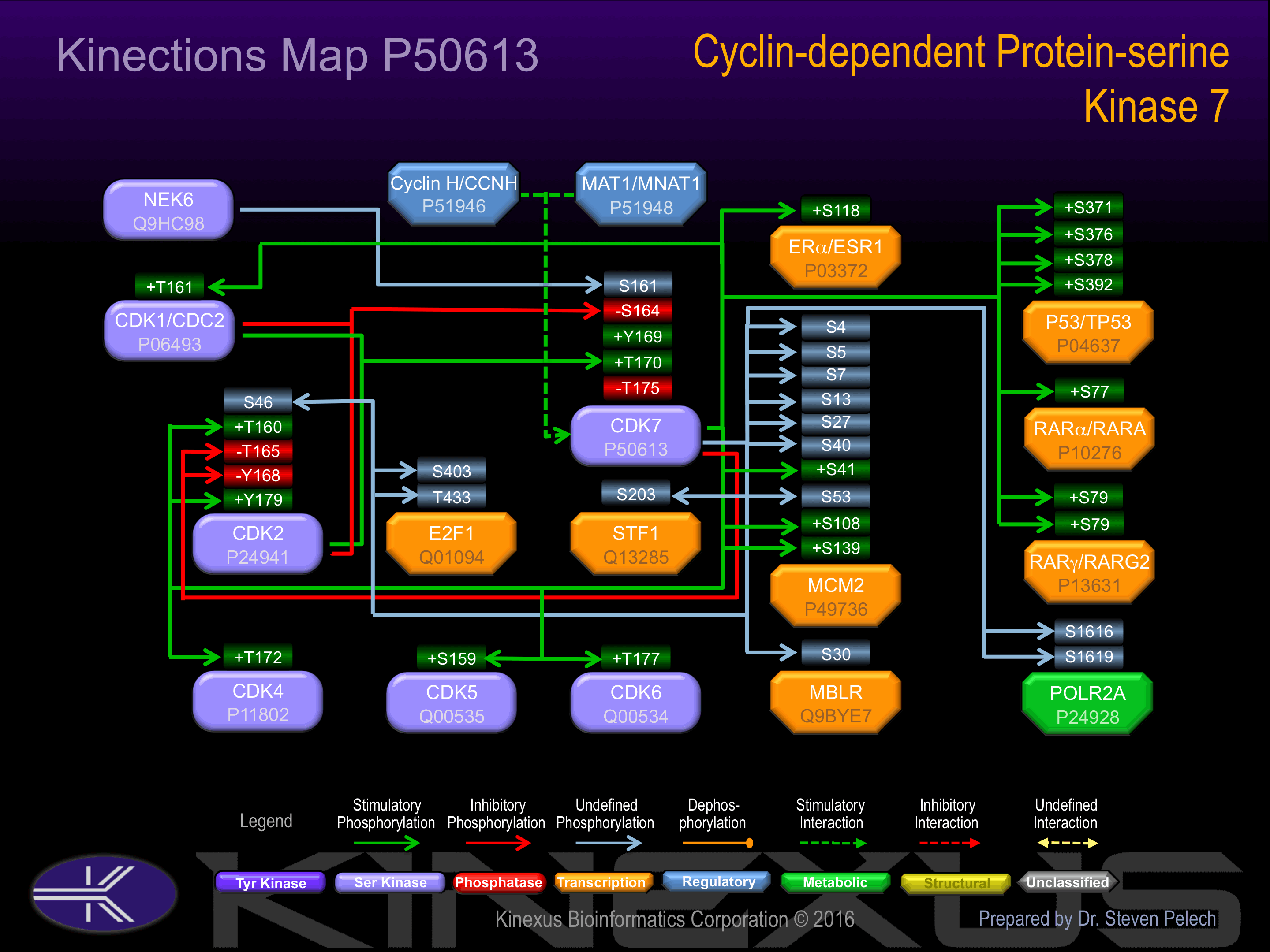
Phosphorylation at Thr-170 is required for activity.
Inhibition:
Phosphorylation of Ser-164 during mitosis inactivates the enzyme.
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
| CDK1 (CDC2) | P06493 | T161 | GIPIRVYTHEVVTLW | + |
| CDK2 | P24941 | S46 | TETEGVPSTAIREIS | |
| CDK2 | P24941 | T160 | GVPVRTYTHEVVTLW | + |
| CDK2 | P24941 | T165 | TYTHEVVTLWYRAPE | - |
| CDK2 | P24941 | Y168 | HEVVTLWYRAPEILL | - |
| CDK2 | P24941 | Y179 | EILLGCKYYSTAVDI | + |
| CDK4 | P11802 | T172 | YSYQMALTPVVVTLW | + |
| CDK5 | Q00535 | S159 | GIPVRCYSAEVVTLW | + |
| CDK6 | Q00534 | T177 | YSFQMALTSVVVTLW | + |
| E2F1 | Q01094 | S403 | PEEFISLSPPHEALD | |
| E2F1 | Q01094 | T433 | DCDFGDLTPLDF___ | |
| ERa (ESR1) | P03372 | S118 | LHPPPQLSPFLQPHG | + |
| MBLR | Q9BYE7 | S30 | LPPPPPVSPPALTPA | |
| MCM2 | P49736 | S108 | DVEELTASQREAAER | + |
| MCM2 | P49736 | S13 | ESFTMASSPAQRRRG | ? |
| MCM2 | P49736 | S139 | RRGLLYDSDEEDEER | + |
| MCM2 | P49736 | S27 | GNDPLTSSPGRSSRR | ? |
| MCM2 | P49736 | S4 | ____MAESSESFTMA | |
| MCM2 | P49736 | S40 | RRTDALTSSPGRDLP | ? |
| MCM2 | P49736 | S41 | RTDALTSSPGRDLPP | + |
| MCM2 | P49736 | S5 | ___MAESSESFTMAS | |
| MCM2 | P49736 | S53 | LPPFEDESEGLLGTE | ? |
| MCM2 | P49736 | S7 | _MAESSESFTMASSP | |
| MCM2 | P49736 | T59 | ESEGLLGTEGPLEEE | |
| p53 | P04637 | S33 | LPENNVLSPLPSQAM | + |
| p53 | P04637 | S371 | AHSSHLKSKKGQSTS | + |
| p53 | P04637 | S376 | LKSKKGQSTSRHKKL | + |
| p53 | P04637 | S378 | SKKGQSTSRHKKLMF | + |
| p53 | P04637 | S392 | FKTEGPDSD______ | + |
| POLR2A | P24928 | S1616 | TPQSPSYSPTSPSYS | + |
| POLR2A | P24928 | S1619 | SPSYSPTSPSYSPTS | + |
| RARA | P10276 | S77 | EIVPSPPSPPPLPRI | + |
| RARG2 | P13631 | S77 | SEEMVPSSPSPPPPP | + |
| RARG2 | P13631 | S79 | EMVPSSPSPPPPPRV | + |
| STF1 | Q13285 | S203 | EYPEPYASPPQPGLP |
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
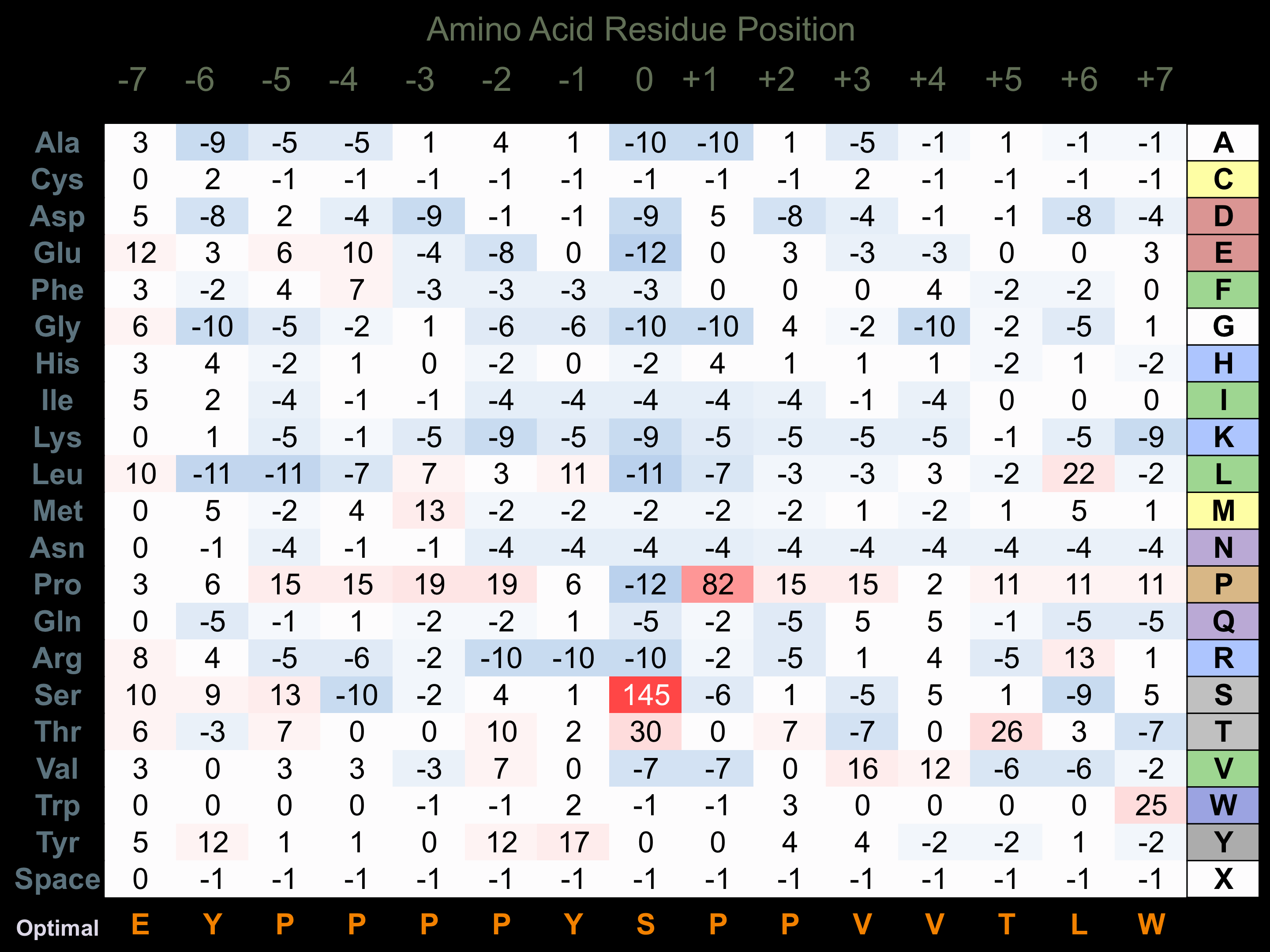
Matrix Type:
Experimentally derived from alignment of 40 known protein substrate phosphosites.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Skin disorders
Specific Diseases (Non-cancerous):
Photosensitive trichothiodystrophy
Comments:
Trichothiodystrophy is associated with mutations in the XPD and XPB subunits of the DNA repair associated transcription factor TFIIH. Along with p63 and p44, CDK7 is a component is the TFIIH complex. Reduced expression levels of all components of the TFIIH complex, including CDK7, are linked to the trichothiodystrophy phenotype. Photosensitive trichothiodystrophy (TTD) is a rare inherited disease characterized by abnormally brittle and sparse hair. This disease is thought to be caused by a sulphur deficiency, as sulphur is used in the normal structure of hair and gives it strength.,The pharmacological modulation of CDK7 activity with the CDK7-specific inhibitor THZ1 has been suggested as a potential approach to identify and treat different tumour types that require transcription for sustaining the oncogenic state, due to the role of CDK7 as an enzymatic cofactor in the transcriptional machinery. Photosensitive trichothiodystrophy (TTD) is a rare inherited disease characterized by abnormally brittle and sparse hair. This disease is thought to be caused by a sulphur deficiency, as sulphur is used in the normal structure of hair and gives it strength.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Barrett's esophagus epithelial metaplasia (%CFC= -47, p<0.021); Bladder carcinomas (%CFC= +173, p<0.0002); Cervical cancer (%CFC= +81, p<0.002); Colon mucosal cell adenomas (%CFC= +89, p<0.0001); Large B-cell lymphomas (%CFC= +85, p<0.019); Malignant pleural mesotheliomas (MPM) tumours (%CFC= +198, p<0.0001); Ovary adenocarcinomas (%CFC= -56, p<0.0002); and Skin squamous cell carcinomas (%CFC= +75, p<0.009). The COSMIC website notes an up-regulated expression score for CDK7 in diverse human cancers of 389, which is 0.8-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 77 for this protein kinase in human cancers was 1.3-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.05 % in 25357 diverse cancer specimens. This rate is only -33 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Frequency of Mutated Sites:
None > 2 in 20,640 cancer specimens
Comments:
No deletions, insertions or complex mutations are noted on the COSMIC website.