Nomenclature
Short Name:
PAK4
Full Name:
Serine-threonine-protein kinase PAK 4
Alias:
- EC 2.7.11.1
- P21-activated kinase 4
Classification
Type:
Protein-serine/threonine kinase
Group:
STE
Family:
STE20
SubFamily:
PAKB
Specific Links
Structure
Mol. Mass (Da):
64,072
# Amino Acids:
591
# mRNA Isoforms:
4
mRNA Isoforms:
64,072 Da (591 AA; O96013); 54,940 Da (501 AA; O96013-4); 48,268 Da (438 AA; O96013-3); 47,924 Da (426 AA; O96013-2
4D Structure:
Interacts with FGFR2 and GRB2 By similarity. Interacts tightly with GTP-bound but not GDP-bound CDC42/p21 and weakly with RAC1. Interacts with its substrate ARHGEF2
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment

Domain Distribution:
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Acetylated:
K455, K467.
Methylated:
K78.
Serine phosphorylated:
S41, S97, S99, S104, S142, S148, S167, S181, S195, S258, S267, S291, S445, S474+, S488.
Threonine phosphorylated:
T187, T207, T277, T478-.
Tyrosine phosphorylated:
Y208+, Y480-.
Ubiquitinated:
K31.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 70
70
1089
42
1425
 7
7
104
20
77
 3
3
52
3
58
 26
26
396
116
581
 53
53
821
35
728
 4
4
56
111
44
 16
16
244
43
462
 100
100
1552
41
2429
 39
39
606
24
504
 11
11
177
101
200
 8
8
118
28
100
 53
53
830
211
704
 2
2
32
36
25
 6
6
93
15
69
 13
13
198
19
200
 8
8
123
22
87
 22
22
334
122
1571
 18
18
276
14
219
 3
3
51
111
48
 35
35
540
162
605
 9
9
133
16
87
 7
7
101
22
95
 5
5
74
6
65
 10
10
148
14
146
 6
6
90
16
71
 71
71
1099
66
1584
 5
5
72
39
64
 10
10
148
14
101
 7
7
113
14
87
 18
18
285
42
219
 32
32
490
42
461
 47
47
736
51
808
 55
55
861
83
1386
 51
51
788
78
668
 3
3
52
48
46
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 49.3
49.3
59.7
0 91.7
91.7
92.4
97 -
-
-
94 -
-
-
99 95.1
95.1
97
96 -
-
-
- 92.8
92.8
94.4
93 34
34
52.5
93 -
-
-
- 54.3
54.3
63.4
- 65.7
65.7
71.2
80 64.8
64.8
72.6
76 65
65
72.1
72.5 -
-
-
- 50.1
50.1
61.5
69 50
50
62.5
- 32.3
32.3
47.5
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 26.9
26.9
41.2
- 28.6
28.6
41.4
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | CDC42 - P60953 |
| 2 | RAC3 - P60763 |
| 3 | ITGB5 - P18084 |
| 4 | YWHAZ - P63104 |
| 5 | YWHAH - Q04917 |
| 6 | YWHAG - P61981 |
| 7 | PAK1 - Q13153 |
Regulation
Activation:
Activated by binding the small G protein CDC42. Binding of CDC42 to the autoregulatory region releases monomers from the autoinhibited dimer, enables phosphorylation of Ser-474 in the kinase activation loop and allows the kinase domain to adopt an active structure.
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Upstream Kinases
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Kinase Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Protein Kinase Specificity
Matrix of observed frequency (%) of amino acids in aligned protein substrate phosphosites
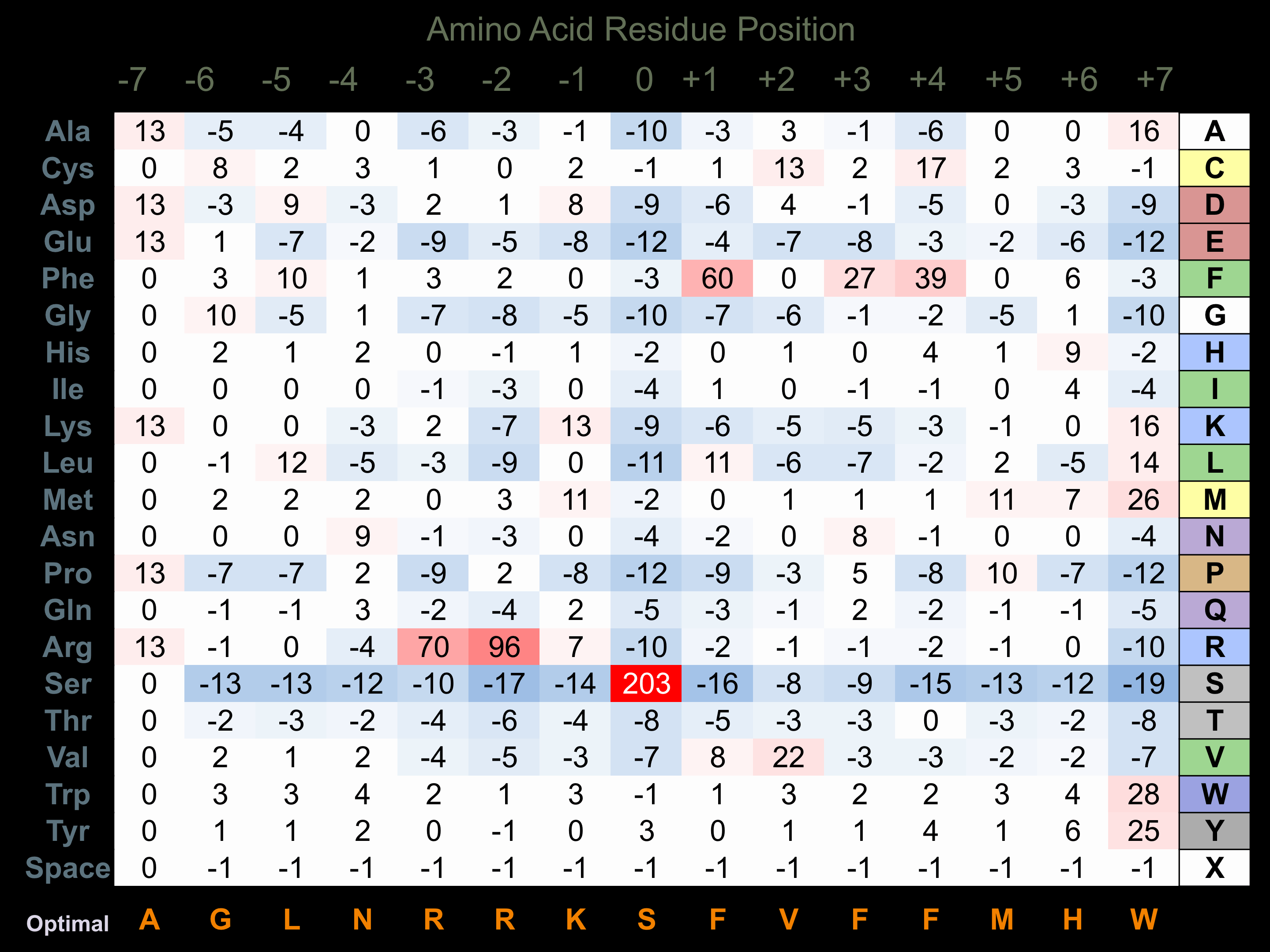
Matrix Type:
Experimentally derived from alignment of 6 known protein substrate phosphosites and 27 peptides phosphorylated by recombinant PAK4 in vitro tested in-house by Kinexus.
Domain #:
1
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
General Disease Association:
Cancer
Specific Cancer Types:
Breast adenomas
Comments:
PAK4 mutations are associated with increased carcinoma cell motility and influenced inhibitor sensitivity. The protein kinase was found to be necessary for oncogenic transformation of breast cancer cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain oligodendrogliomas (%CFC= +168, p<0.062); and Oral squamous cell carcinomas (OSCC) (%CFC= +54, p<0.002). The COSMIC website notes an up-regulated expression score for PAK4 in diverse human cancers of 791, which is 1.7-fold of the average score of 462 for the human protein kinases. The down-regulated expression score of 129 for this protein kinase in human cancers was 2.2-fold of the average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice support a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25409 diverse cancer specimens. This rate is only -15 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.51 % in 1128 large intestine cancers tested; 0.36 % in 807 skin cancers tested.
Frequency of Mutated Sites:
Most frequent mutations with the number of reports indicated in brackets: R252* (4).
Comments:
Only 7 deletions, no insertions or complex mutations are noted on the COSMIC website.

