Nomenclature
Short Name:
PDHK1
Full Name:
[Pyruvate dehydrogenase [lipoamide]] kinase isozyme 1, mitochondrial
Alias:
- Kinase Pyruvate dehydrogenase kinase 1
- Pyruvate dehydrogenase [lipoamide] kinase isozyme 1, mitochondrial precursor
- Pyruvate dehydrogenase kinase isoform 1
- Pyruvate dehydrogenase kinase, isozyme 1
- PDK1
Classification
Type:
Protein-serine/threonine kinase
Group:
Atypical
Family:
PDHK
SubFamily:
NA
Specific Links
Structure
Mol. Mass (Da):
49,244
# Amino Acids:
436
# mRNA Isoforms:
2
mRNA Isoforms:
51,623 Da (456 AA; Q15118-2); 49,244 Da (436 AA; Q15118)
4D Structure:
NA
1D Structure:
3D Image (rendered using PV Viewer):
PDB ID
Subfamily Alignment
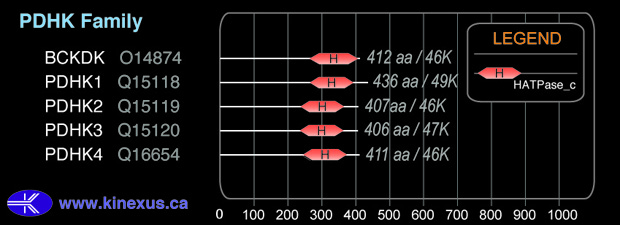
Domain Distribution:
| Start | End | Domain |
|---|---|---|
| 268 | 393 | HATPase_c |
| 163 | 393 | Histidine kinase |
| 55 | 221 | BCDHK |
Kinexus Products
Click on entries below for direct links to relevant products from Kinexus for this protein kinase.
hiddentext
Post-translation Modifications
For detailed information on phosphorylation of this kinase go to PhosphoNET
Serine phosphorylated:
S67, S347.
Threonine phosphorylated:
T346.
Tyrosine phosphorylated:
Y136, Y243, Y244, Y336, Y357, Y403.
Distribution
Based on gene microarray analysis from the NCBI
Human Tissue Distribution
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
% Max Expression:
Mean Expression:
Number of Samples:
Standard Deviation:
 35
35
1029
22
1086
 2
2
50
11
88
 2
2
71
12
79
 35
35
1020
88
2753
 31
31
897
21
611
 22
22
654
54
1971
 7
7
204
29
356
 31
31
901
40
2091
 8
8
247
10
241
 4
4
129
84
215
 1.4
1.4
42
29
64
 15
15
432
126
565
 5
5
147
23
191
 0.3
0.3
10
8
5
 1
1
29
14
34
 6
6
176
13
211
 1.3
1.3
37
236
40
 3
3
73
19
92
 2
2
50
72
58
 28
28
807
84
772
 1.3
1.3
38
25
44
 3
3
80
27
93
 5
5
135
20
153
 5
5
137
19
126
 3
3
79
25
88
 66
66
1927
59
3637
 3
3
86
26
118
 3
3
86
19
103
 1.2
1.2
34
18
46
 2
2
61
28
48
 28
28
822
18
515
 100
100
2909
26
5386
 12
12
345
66
514
 27
27
793
57
726
 14
14
413
35
300
Evolution
Species Conservation
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
PhosphoNET % Identity:
PhosphoNET % Similarity:
Homologene %
Identity:
 100
100
100
100 98.8
98.8
99.1
99 81
81
84.6
- -
-
-
95 -
-
-
99 62
62
78.8
97 -
-
-
- 91.3
91.3
94
92 92.2
92.2
94.5
93 -
-
-
- 82.3
82.3
86.2
- 83
83
89
89 78.2
78.2
87.8
83 72.3
72.3
83
78.5 -
-
-
- 51.8
51.8
69
- -
-
-
- 45.9
45.9
64.9
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- -
-
-
- 20.6
20.6
41.8
- -
-
-
-
For a wider analysis go to PhosphoNET Evolution in PhosphoNET
Binding Proteins
Examples of known interacting proteins
hiddentext
| No. | Name – UniProt ID |
|---|---|
| 1 | SGK3 - Q96BR1 |
| 2 | PDK2 - Q15119 |
| 3 | PKN2 - Q16513 |
| 4 | RPS6KB1 - P23443 |
| 5 | EPAS1 - Q99814 |
| 6 | DLAT - P10515 |
| 7 | PDHA1 - P08559 |
Regulation
Activation:
NA
Inhibition:
NA
Synthesis:
NA
Degradation:
NA
Known Downstream Substrates
For further details on these substrates click on the Substrate Short Name or UniProt ID. Phosphosite Location is hyperlinked to PhosphoNET
predictions.
Based on in vitro and/or in vivo phosphorylation data
| Substrate Short Name | UniProt ID (Human) | Phosphosite Location | Phosphosite Sequence | Effect of Phosphorylation |
|---|
Inhibitors
For further details on these inhibitors click on the Compound Name and enter it into DrugKiNET or click on the ID's
Based on in vitro and/or in vivo phosphorylation data
| Compound Name | KD, Ki or IC50 (nM) | PubChem ID | ChEMBL ID | PubMed ID |
|---|
Disease Linkage
Comments:
PDHK1 may be a target for antitumour therapy, since PDK1 prevents apoptosis in these cancerous cells.
Gene Expression in Cancers:
TranscriptoNET (www.transcriptonet.ca) analysis with mRNA expression data retrieved from the National Center for Biotechnology Information's Gene Expression Omnibus (GEO) database, which was normalized against 60 abundantly and commonly found proteins, indicated altered expression for this protein kinase as shown here as the percent change from normal tissue controls (%CFC) as supported with the Student T-test in the following types of human cancers: Brain glioblastomas (%CFC= -47, p<0.002); Clear cell renal cell carcinomas (cRCC) (%CFC= +448, p<0.002); Clear cell renal cell carcinomas (cRCC) stage I (%CFC= +3000, p<0.001); Prostate cancer - primary (%CFC= +75, p<0.0001); T-cell prolymphocytic leukemia (%CFC= -48, p<0.01); and Vulvar intraepithelial neoplasia (%CFC= +61, p<0.082). The COSMIC website notes an up-regulated expression score for PDHK1 in diverse human cancers of 451, which is close to the average score of 462 for the human protein kinases. The down-regulated expression score of 2 for this protein kinase in human cancers was 97% lower than average score of 60 for the human protein kinases.
Mutagenesis Experiments:
Insertional mutagenesis studies in mice have not yet revealed a role for this protein kinase in mouse cancer oncogenesis.
Mutation Rate in All Cancers:
Percent mutation rates per 100 amino acids length in human cancers: 0.06 % in 25430 diverse cancer specimens. This rate is only -18 % lower than the average rate of 0.075 % calculated for human protein kinases in general.
Mutation Rate in Specific Cancers:
Highest percent mutation rates per 100 amino acids length in human cancers: 0.34 % in 273 cervix cancers tested; 0.24 % in 866 skin cancers tested; 0.23 % in 1305 large intestine cancers tested; 0.18 % in 1634 lung cancers tested; 0.12 % in 590 stomach cancers tested; 0.11 % in 603 endometrium cancers tested; 0.1 % in 710 oesophagus cancers tested; 0.08 % in 833 ovary cancers tested; 0.06 % in 2083 central nervous system cancers tested; 0.06 % in 1512 liver cancers tested; 0.05 % in 1500 breast cancers tested; 0.04 % in 548 urinary tract cancers tested; 0.03 % in 881 prostate cancers tested; 0.02 % in 2009 haematopoietic and lymphoid cancers tested; 0.02 % in 1276 kidney cancers tested.
Frequency of Mutated Sites:
None > 3 in 20,713 cancer specimens
Comments:
Only 1 insertion, and no deletions or complex mutations are noted in COSMIC website.

